BrainGlobe and napari
Whole brain microscopy analysis
Welcome
Schedule
Morning
- Installation (AT & AF)
- Introduction to BrainGlobe (AT)
- Introduction to napari (AF)
- Napari demo (AF)
- BrainGlobe & napari (AF)
- Whole brain atlas registration & segmentation (AT)
Schedule
Afternoon
- 3D cell detection (AT)
- Analysing silicon probe tracks (AT)
- Analysing bulk fluorescence (AT)
- Using BrainGlobe in your own software (AF)
- Wrap up and Q&A (AT & AF)
Installation
Command line installation
Create a new conda environment and install napari
conda create --name brainglobe python=3.10 -y
conda activate brainglobe
conda install -c conda-forge napari pyqtDouble-check that running
opens a new napari window.
More details
More details about using conda are available at brainglobe.info
Command line installation (alternative)
Create a new conda environment and install napari with pip
Double-check that running
opens a new napari window.
More details
More details about using conda are available at brainglobe.info
Install brainglobe napari plugins
Install all brainglobe tools with
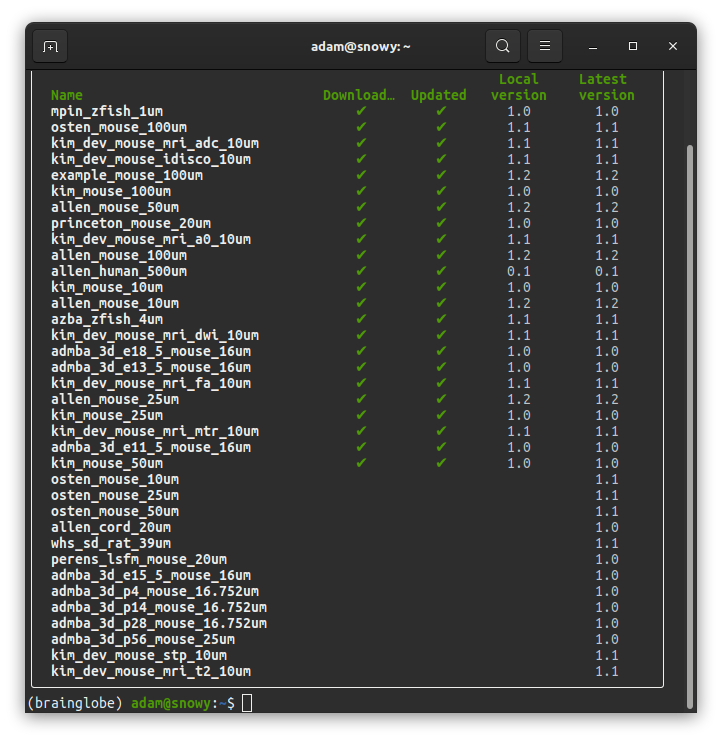
Initial checkup
BrainGlobe
BrainGlobe Initiative
Established 2020 with three aims:
- Develop general-purpose tools to help others build interoperable software
- Develop specialist software for specific analysis and visualisation needs
- Reduce barriers of entry, and facilitate the building of an ecosystem of computational neuroanatomy tools.
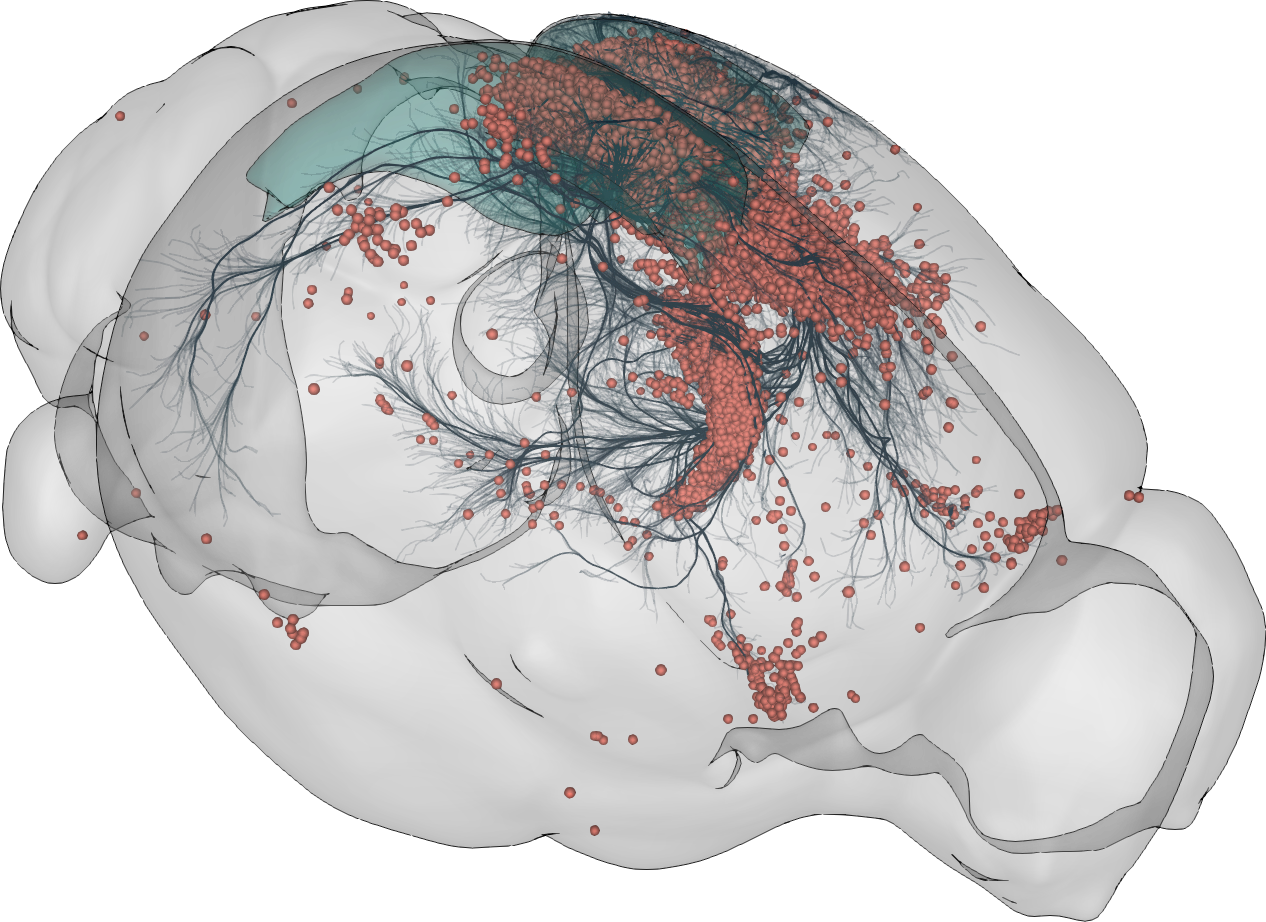
BrainGlobe Atlas API
Initial observation - lots of similar communities working independently
- Model species
- Imaging modality
- Anatomical focus
- Developmental stage
BrainGlobe Atlas API
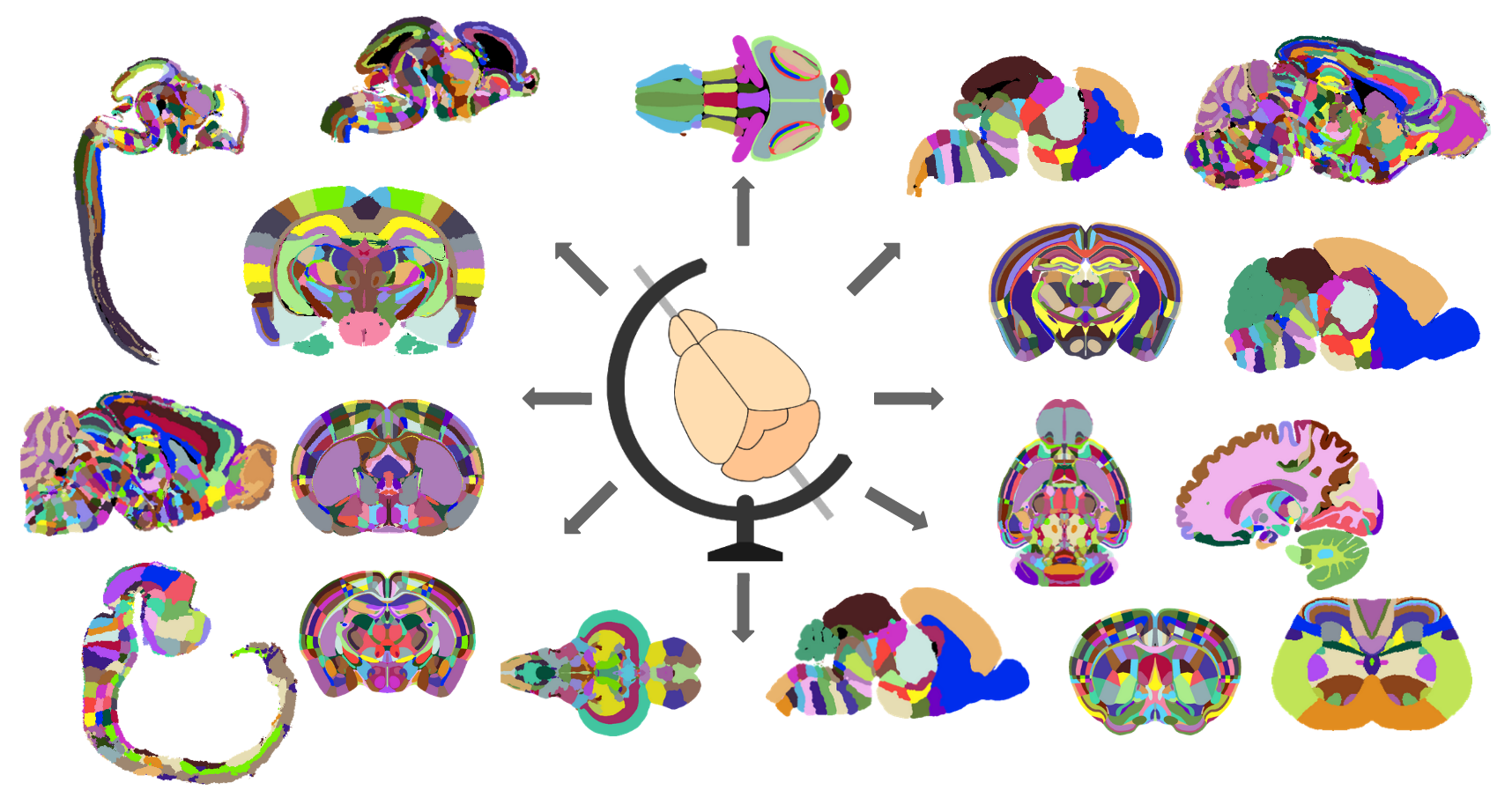
BrainGlobe Atlas API
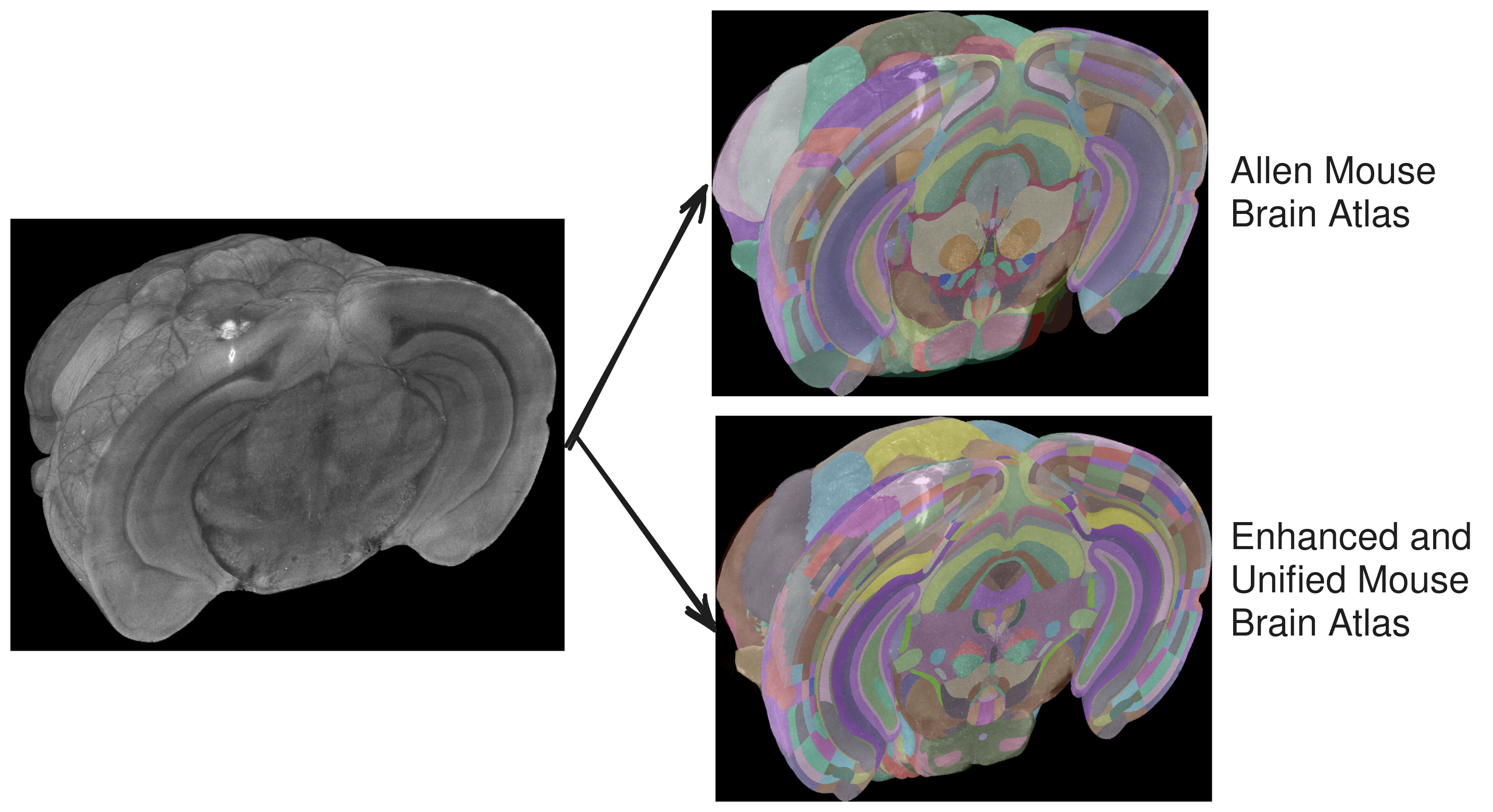
Currently implemented atlases
- Allen Mouse Brain Atlas at 10, 25, 50 and 100 micron resolutions
- Allen Human Brain Atlas at 100 micron resolution
- Max Planck Zebrafish Brain Atlas at 1 micron resolution
- Enhanced and Unified Mouse Brain Atlas at 10, 25, 50 and 100 micron resolutions
- Smoothed version of the Kim et al. mouse reference atlas at 10, 25, 50 and 100 micron resolutions
- Gubra’s LSFM mouse brain atlas at 20 micron resolution
- 3D version of the Allen mouse spinal cord atlas at 20 x 10 x 10 micron resolution
- AZBA: A 3D Adult Zebrafish Brain Atlas at 4 micron resolution
- Waxholm Space atlas of the Sprague Dawley rat brain at 39 micron resolution
- 3D Edge-Aware Refined Atlases Derived from the Allen Developing Mouse Brain Atlases (E13, E15, E18, P4, P14, P28 & P56)
- Princeton Mouse Brain Atlas at 20 micron resolution
- Kim Lab Developmental CCF (P56) at 10 micron resolution with 8 reference images - STP, LSFM (iDISCO) and MRI (a0, adc, dwo, fa, MTR, T2)
BrainGlobe Atlas API
from pprint import pprint
from bg_atlasapi.bg_atlas import BrainGlobeAtlas
atlas = BrainGlobeAtlas("allen_mouse_25um")
# reference image
reference_image = atlas.reference
print(reference_image.shape)
# (528, 320, 456)
# hemispheres image (value 1 in left hemisphere, 2 in right)
hemispheres_image = atlas.hemispheres
print(hemispheres_image.shape)
# (528, 320, 456)
VISp = atlas.structures["VISp"]
pprint(VISp)
# {'acronym': 'VISp',
# 'id': 385,
# 'mesh': None,
# 'mesh_filename': PosixPath('/home/user/.brainglobe/allen_mouse_25um_v0.3/meshes/385.obj')
# 'name': 'Primary visual area',
# 'rgb_triplet': [8, 133, 140],
# 'structure_id_path': [997, 8, 567, 688, 695, 315, 669, 385]}BrainGlobe Atlas API
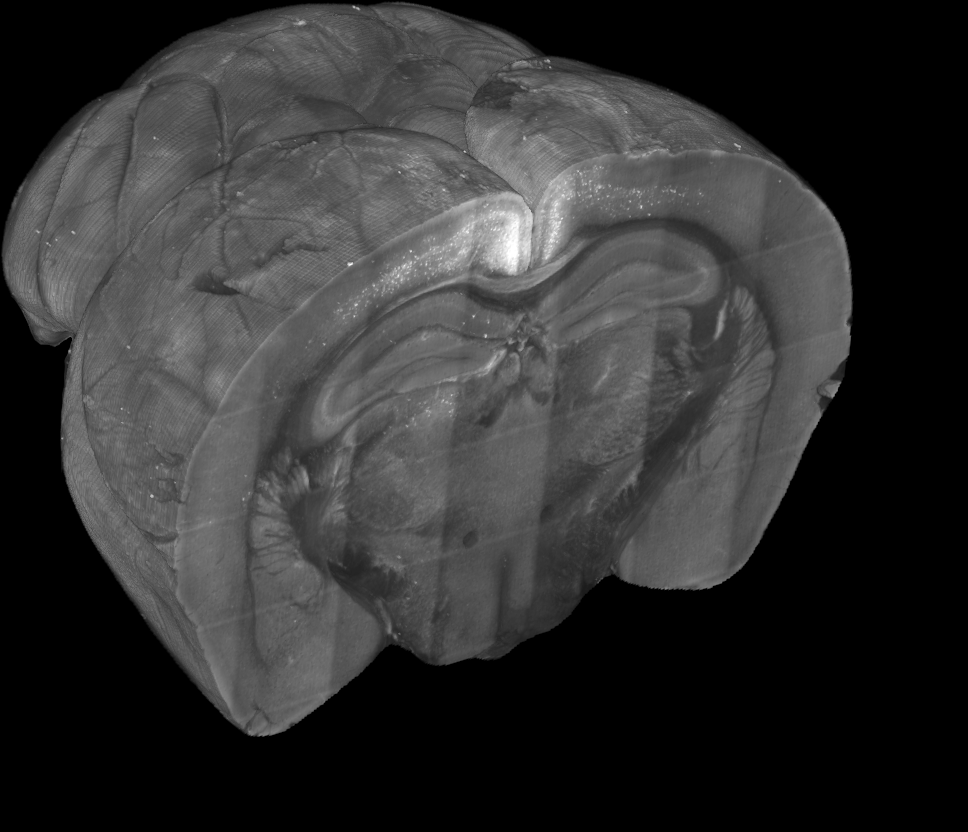
Whole brain microscopy
Serial section two-photon tomography
Fluorescence micro-optical sectioning tomography
Light sheet fluorescence microscopy
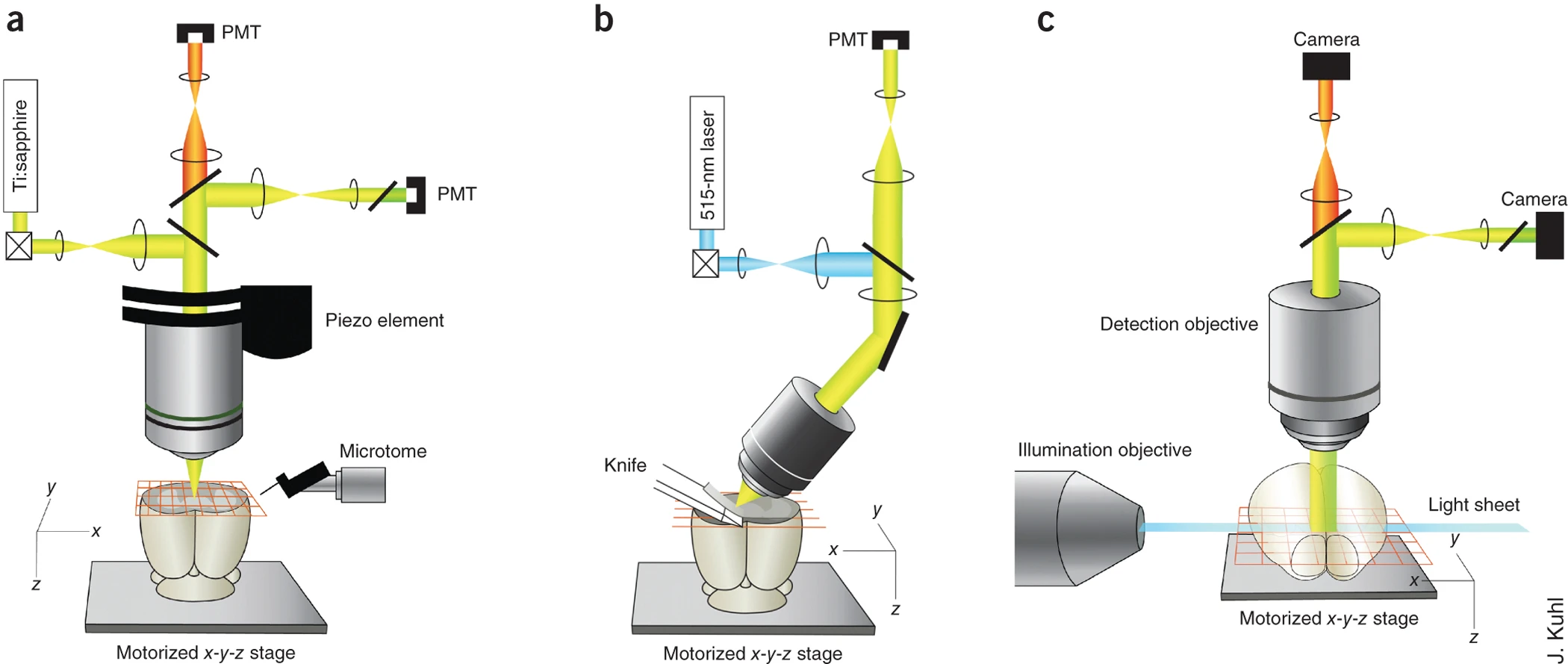
Whole brain microscopy
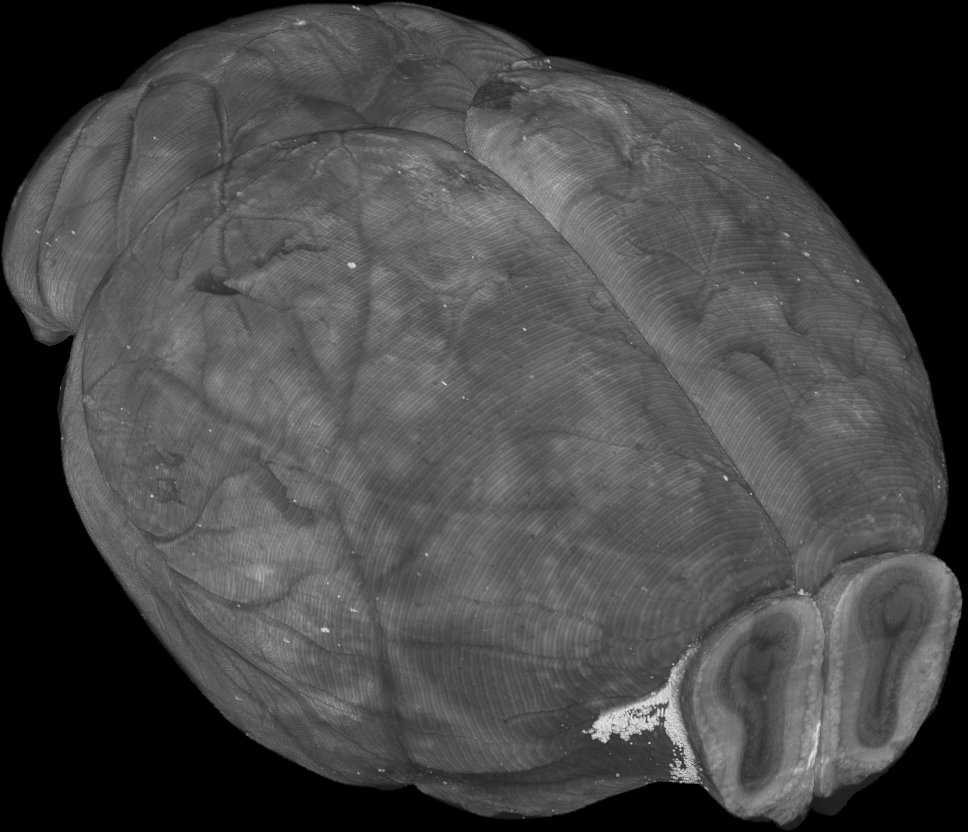
Whole brain microscopy
brainreg
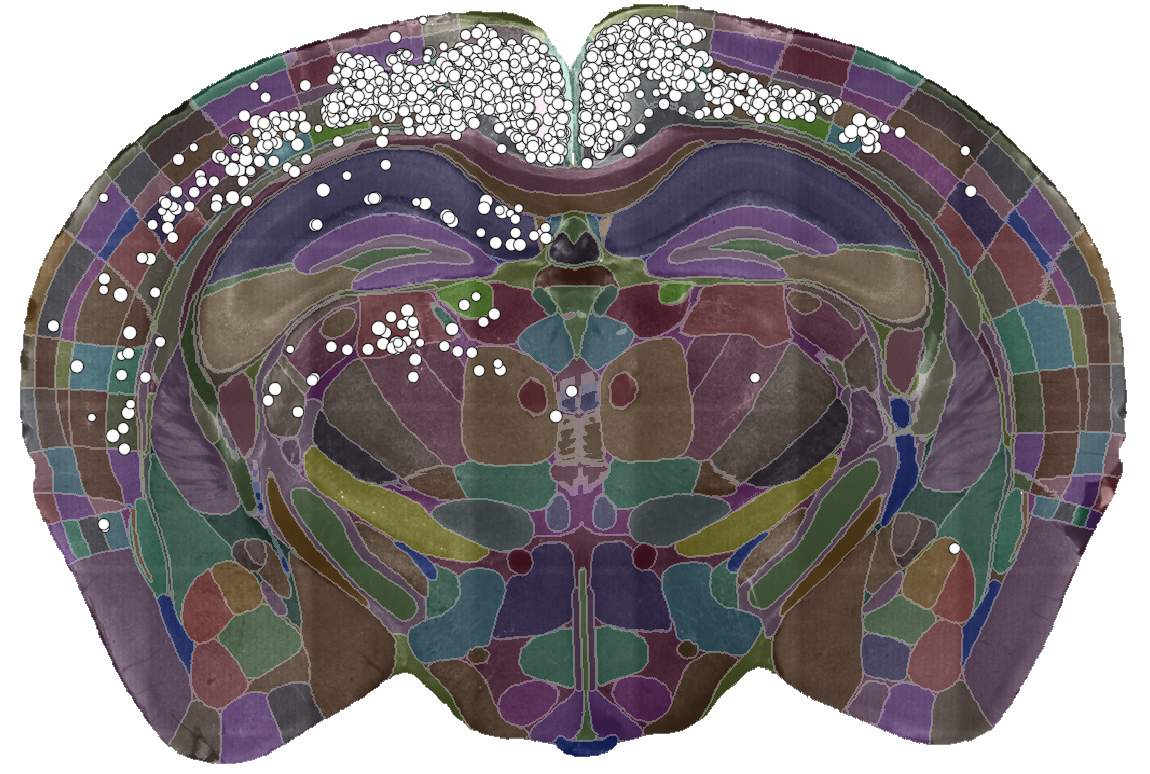
cellfinder
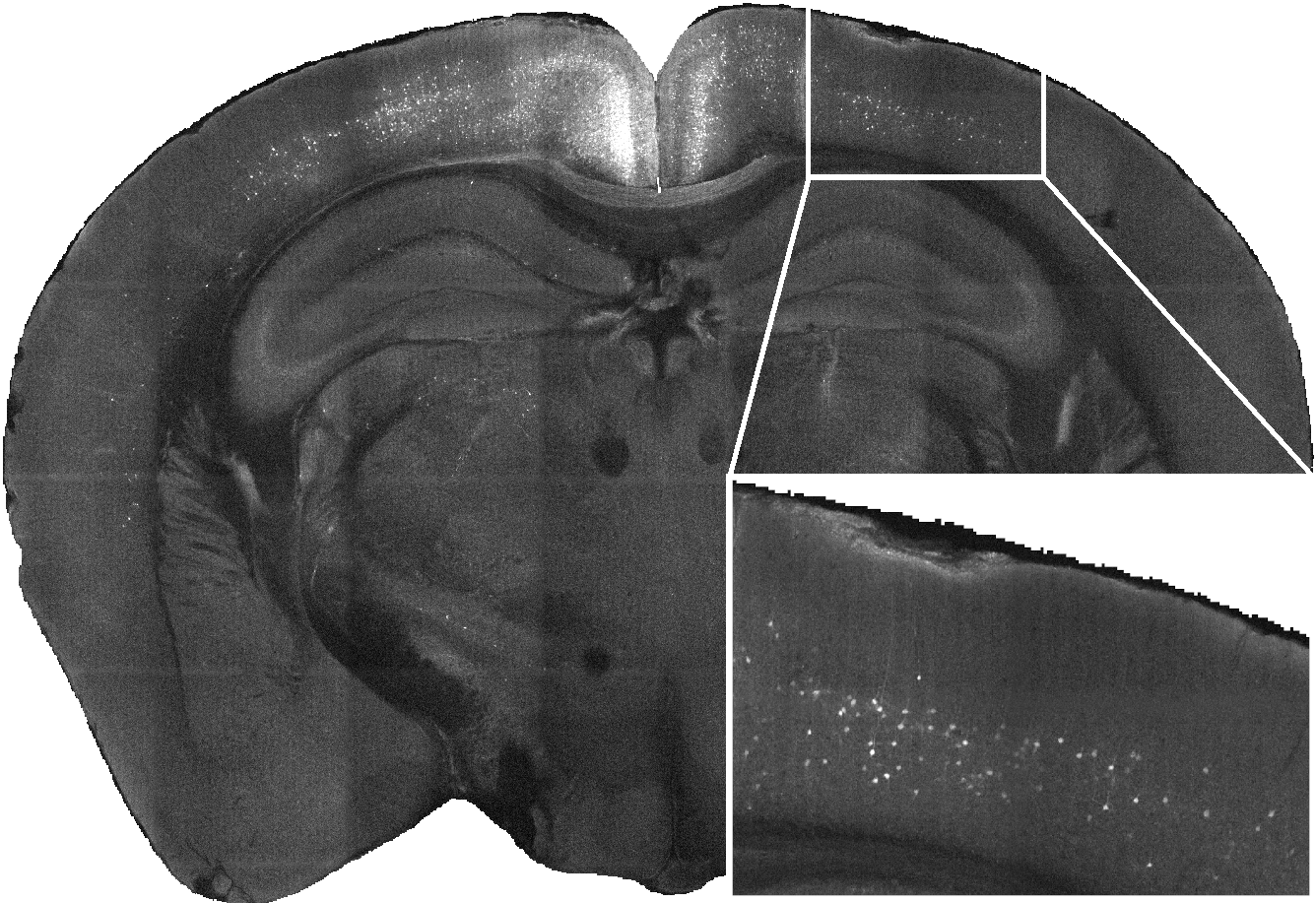
cellfinder
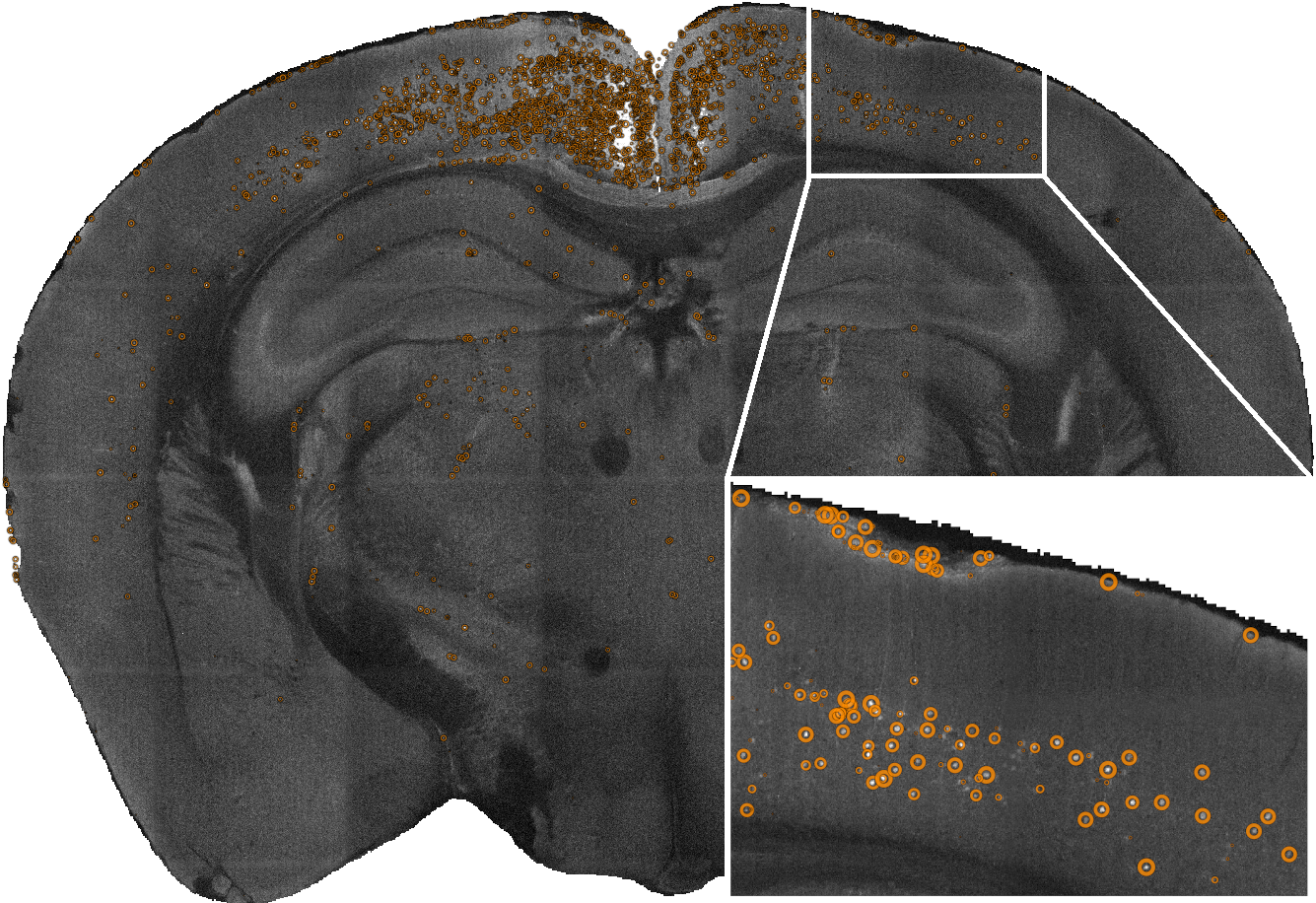
cellfinder
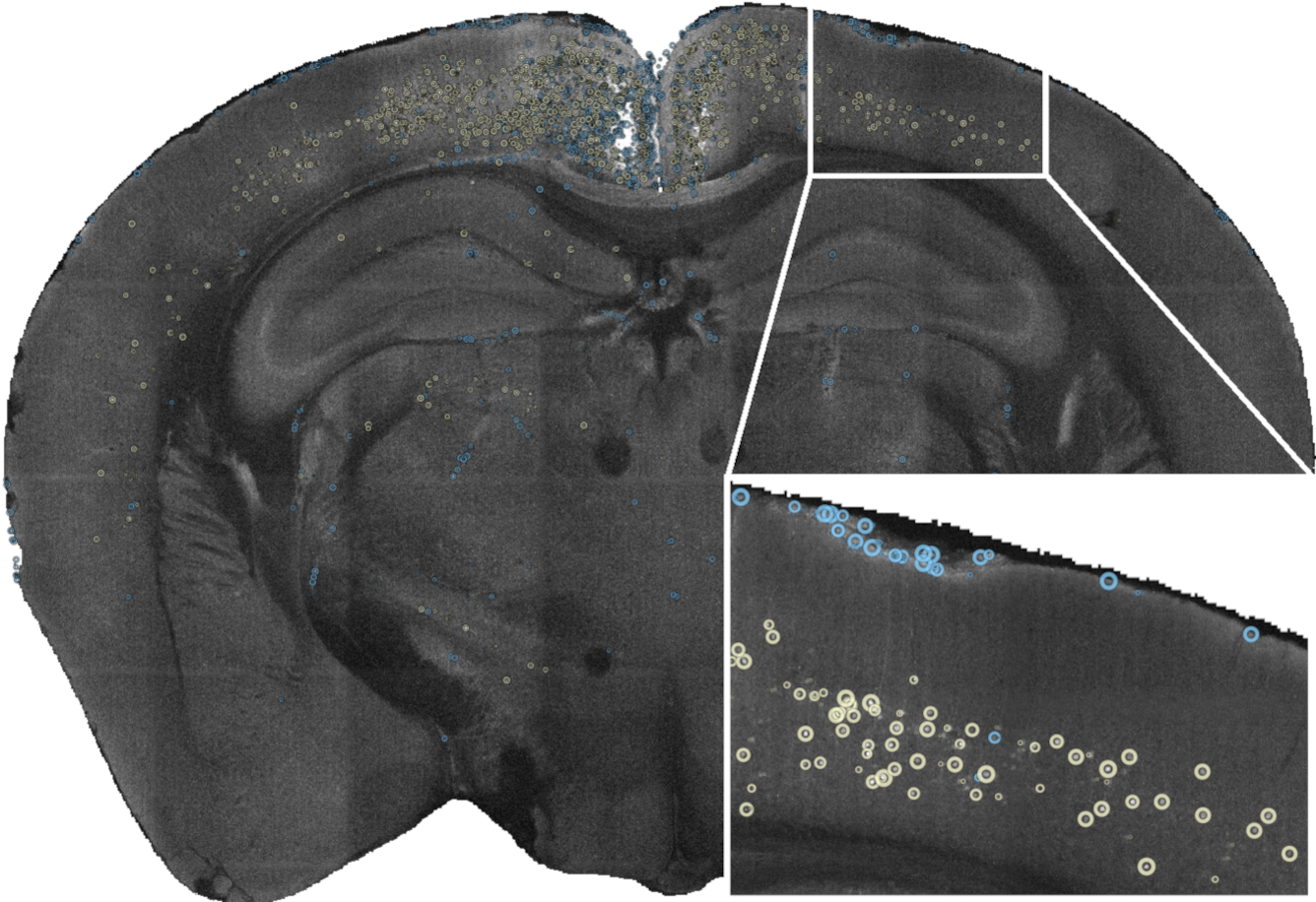
cellfinder
brainglobe-segmentation
brainglobe-segmentation
brainglobe-segmentation
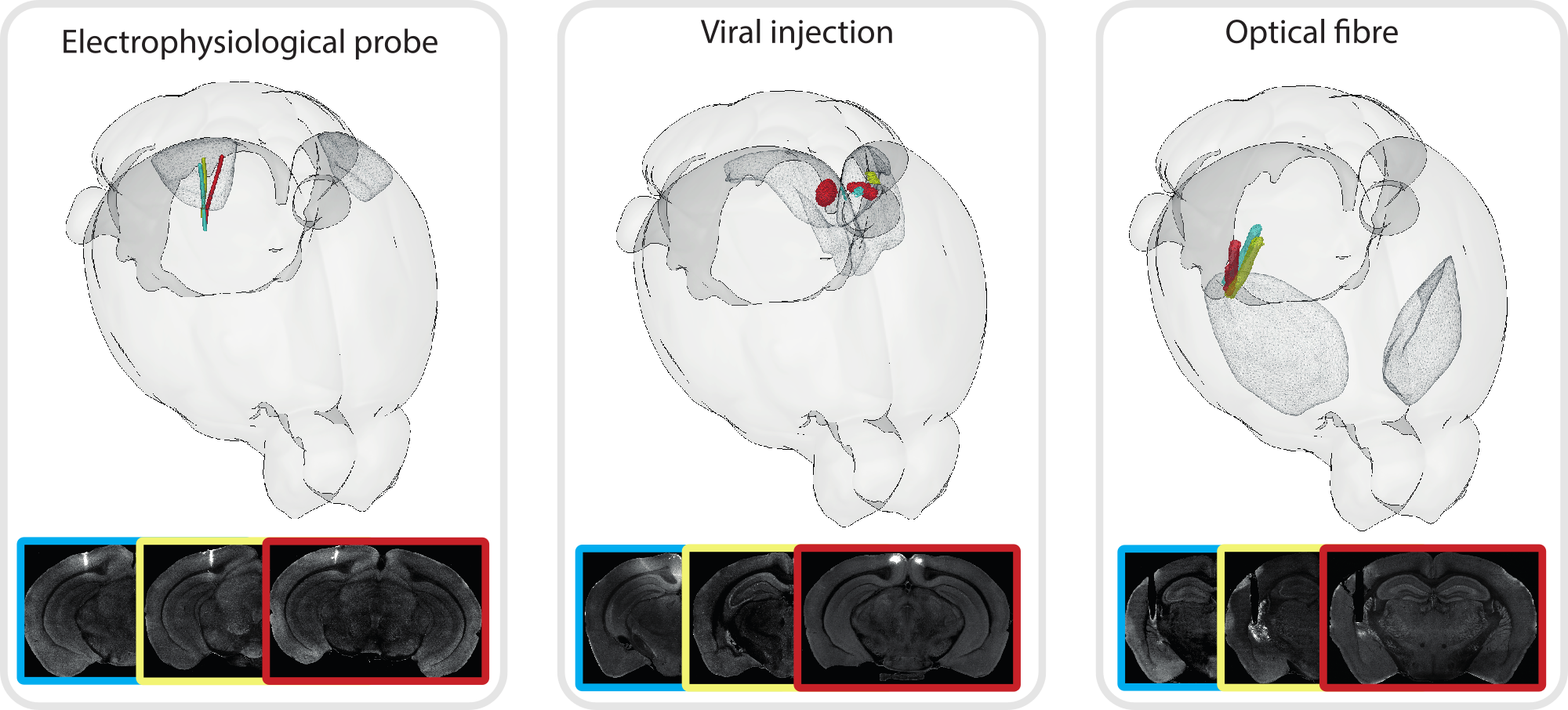
brainrender
brainrender
BrainGlobe website
napari
What does napari look like?
The napari tech stack
Who has used napari before?
- increasing in popularity
- still a bit immature

Basic functionality
On it’s own, napari can:
- open and display multi-dimensional images and overlays
- manipulate these
- install plugins
Why is it good?
It leverages Python well:
- Open source
- Easy access to (scientific) Python (in several ways)
- Plugin installation (and development) easy
- (Relatively) good documentation
- Friendly and diverse community (including designers)
- Funding for plugins in research available
Why is it not-so-good?
The immature ecosystem means:
- things occasionally break (but lots of enthusiastic support available)
- several plugins to do the same thing (please don’t reinvent the wheel - re-use other people’s wheels!)
- installation trickier than e.g. Fiji/ImageJ
The possibilities are endless
Video re-used from Napari-napari under BSD-3 license.
napari demo
napari-demo
- please open
napariand follow allow with the live demo
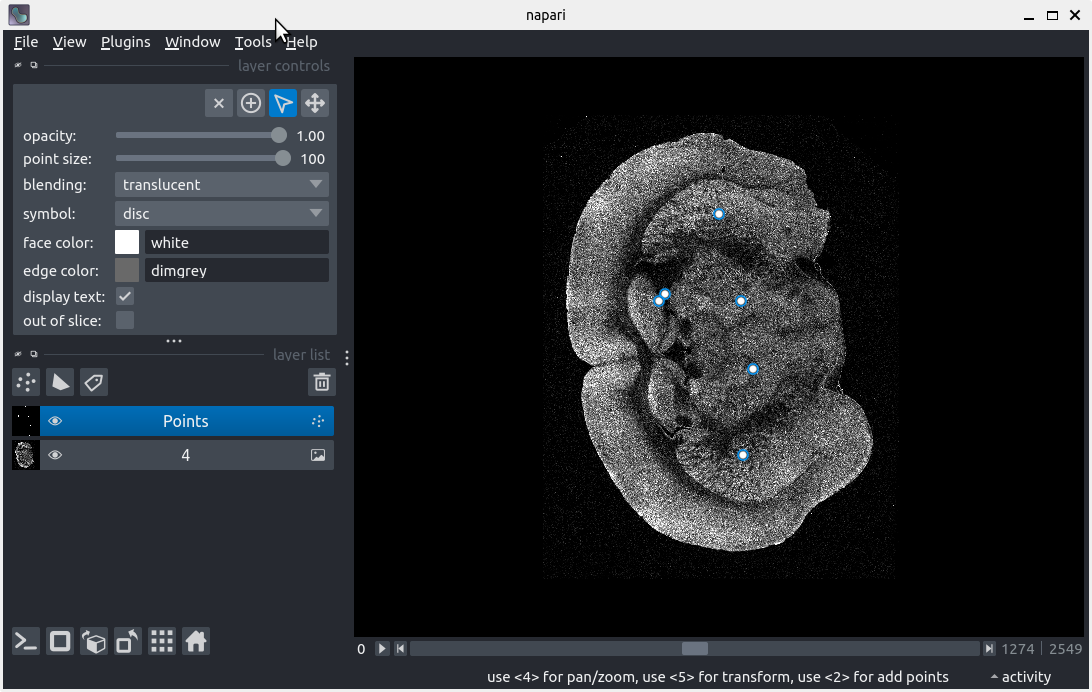
Data exploration
After following along, your screen should look something like:
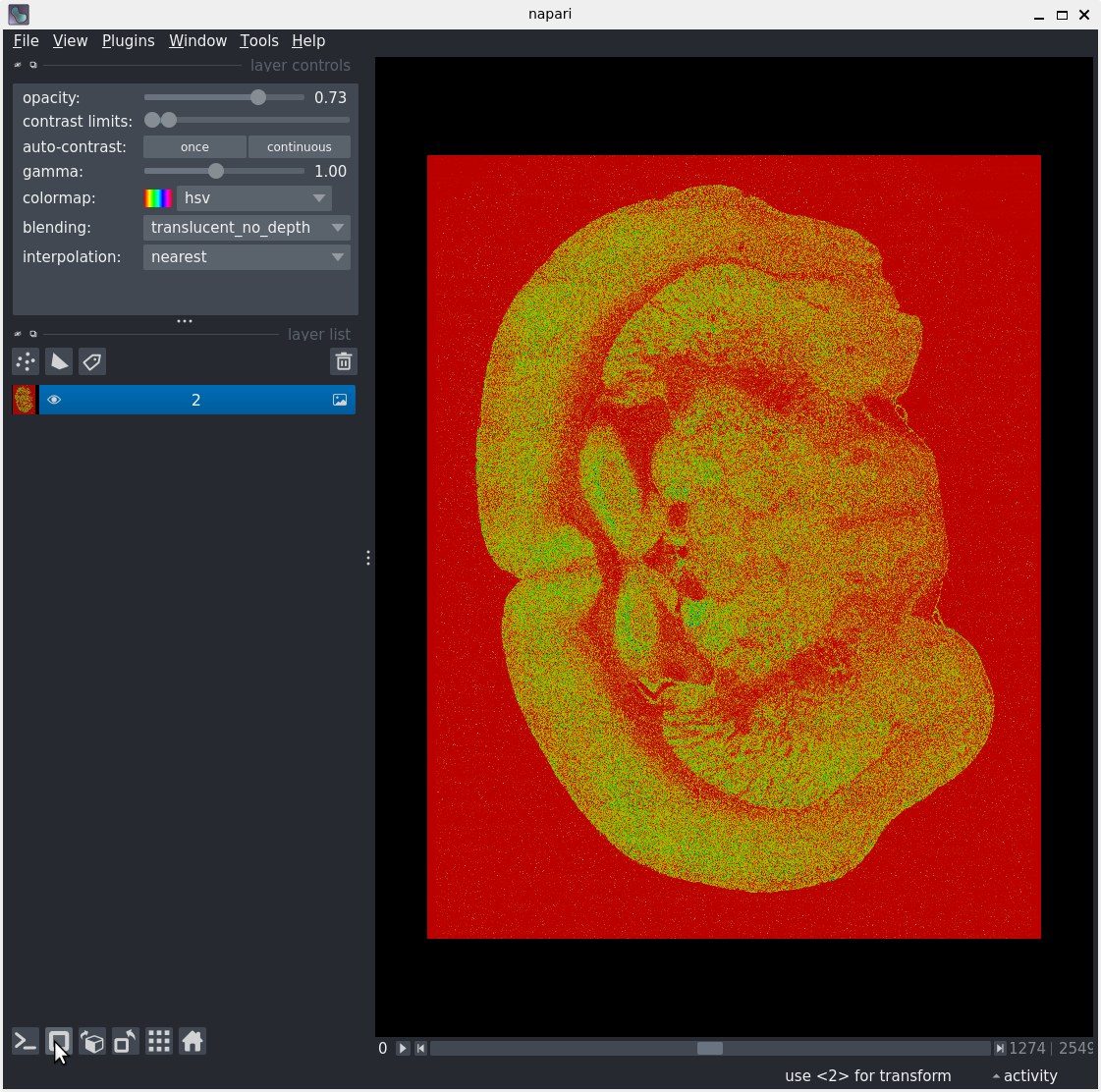
Data exploration

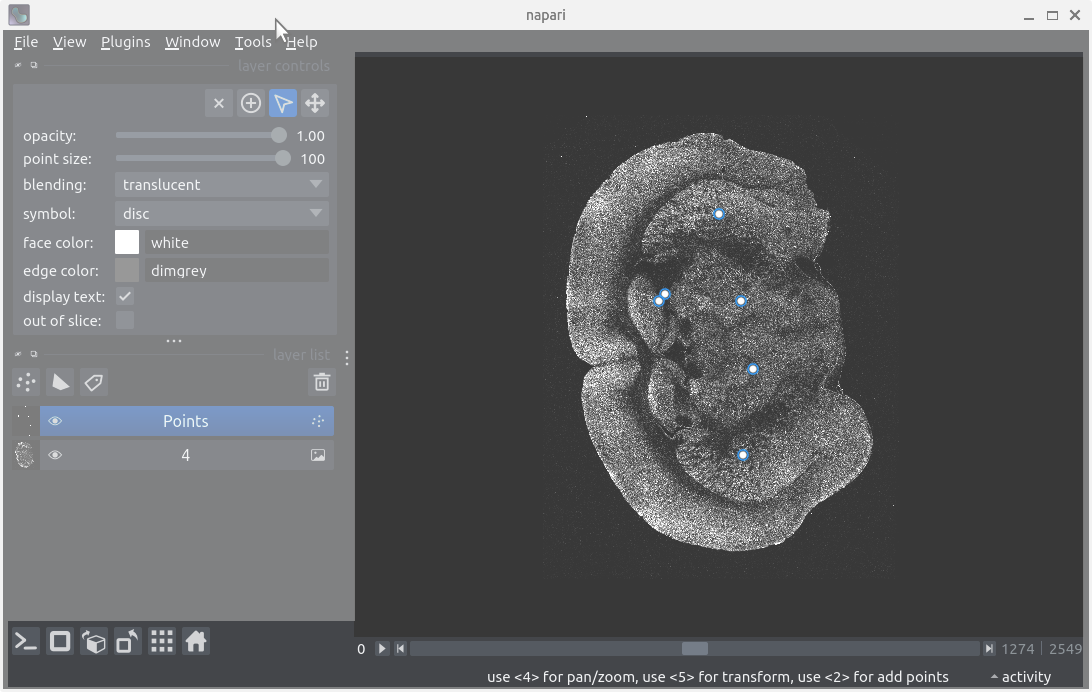
Layer controls
After following along, your screen should look something like:
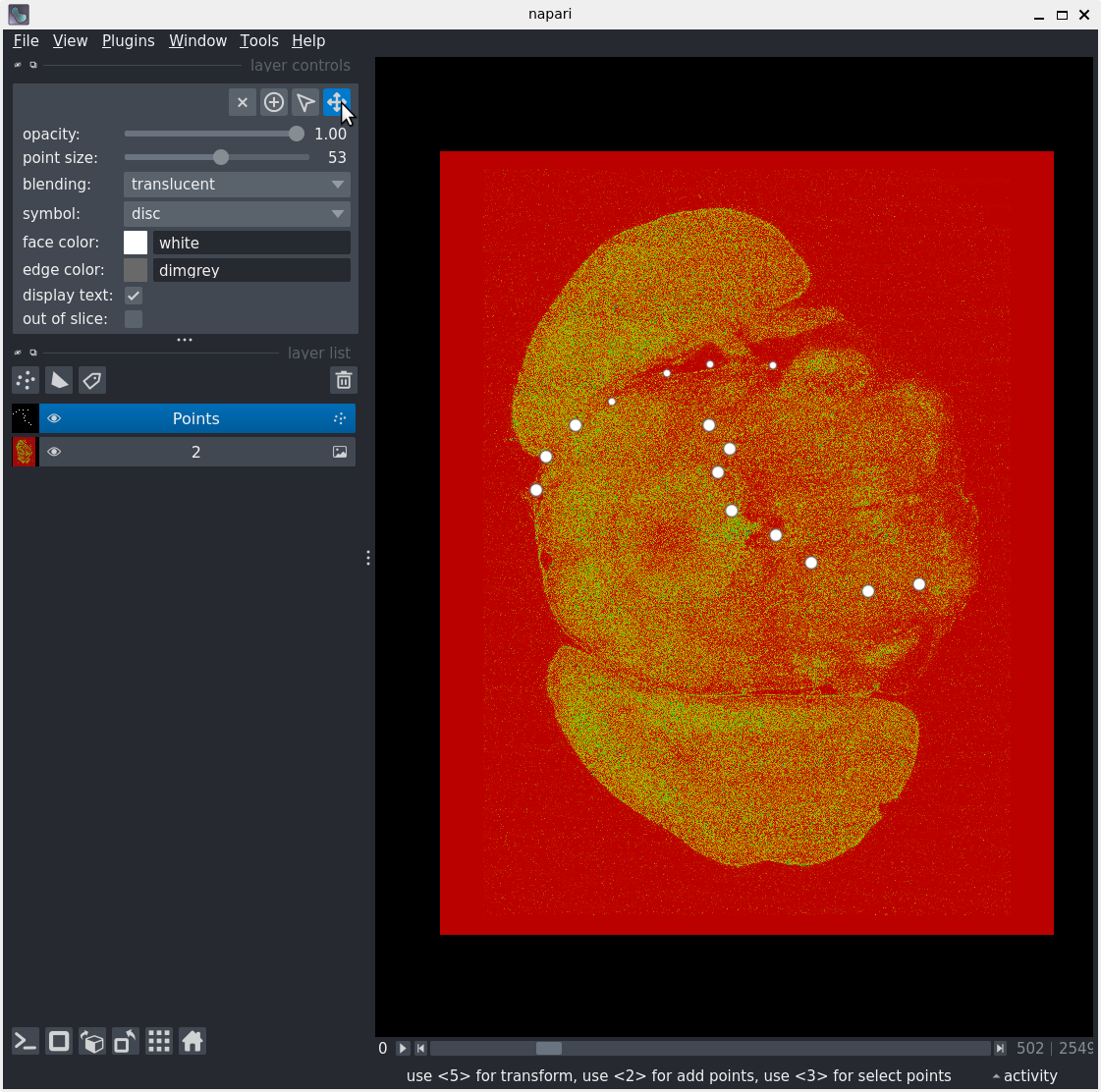
Layer controls

The IPython console
- Navigate to
Window > Console - In the console, run (each line separately)
import napari
from pathlib import Path
from cellfinder_core.tools.IO import read_with_dask
viewer = napari.viewer.current_viewer()
# adapt "path/to/data" to your folder of tiffs
path_to_data = Path("path/to/data")
viewer.open(path_to_data)
print(viewer.layers)
# which of these is quicker?
%timeit -r 3 -n 1 viewer.open(path_to_data)
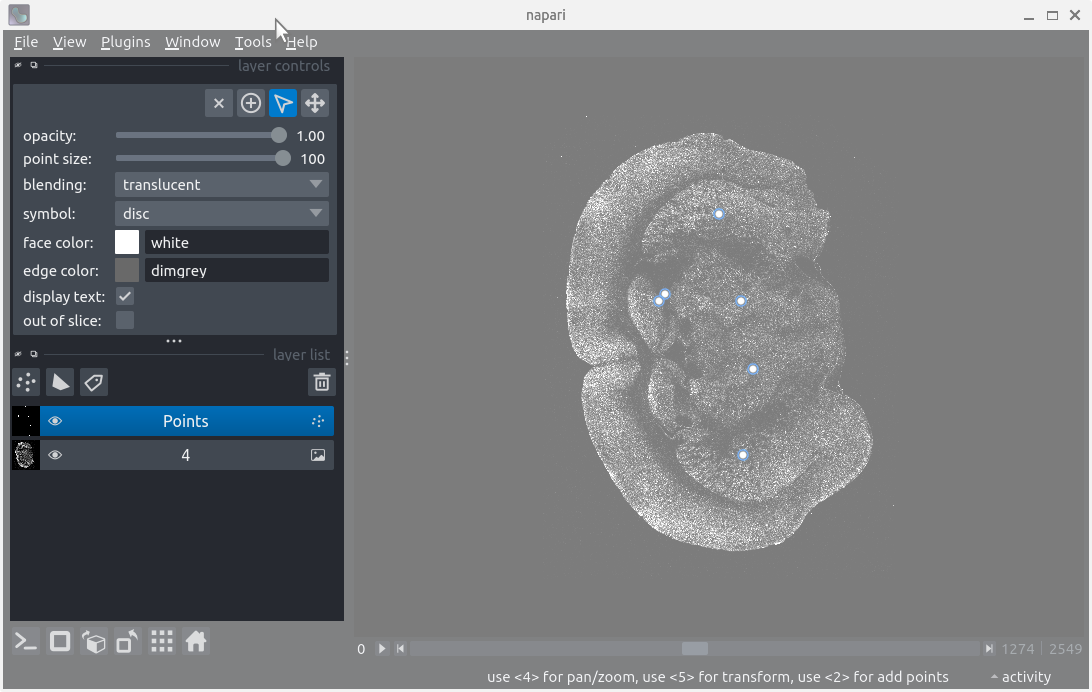
%timeit -r 3 -n 1 viewer.add_image(read_with_dask(str(path_to_data)))The IPython console
After following along, your screen should look something like:
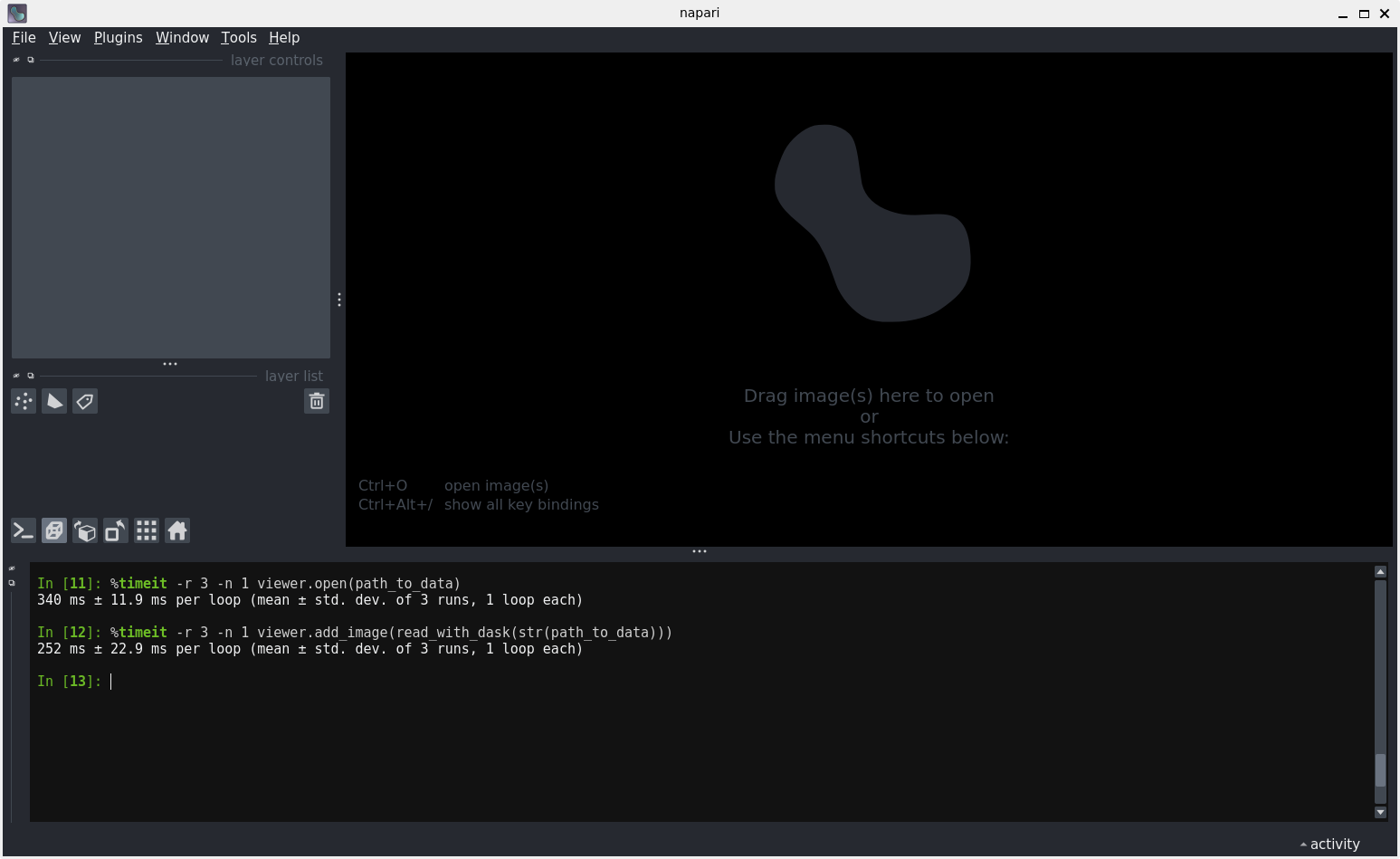
The IPython console

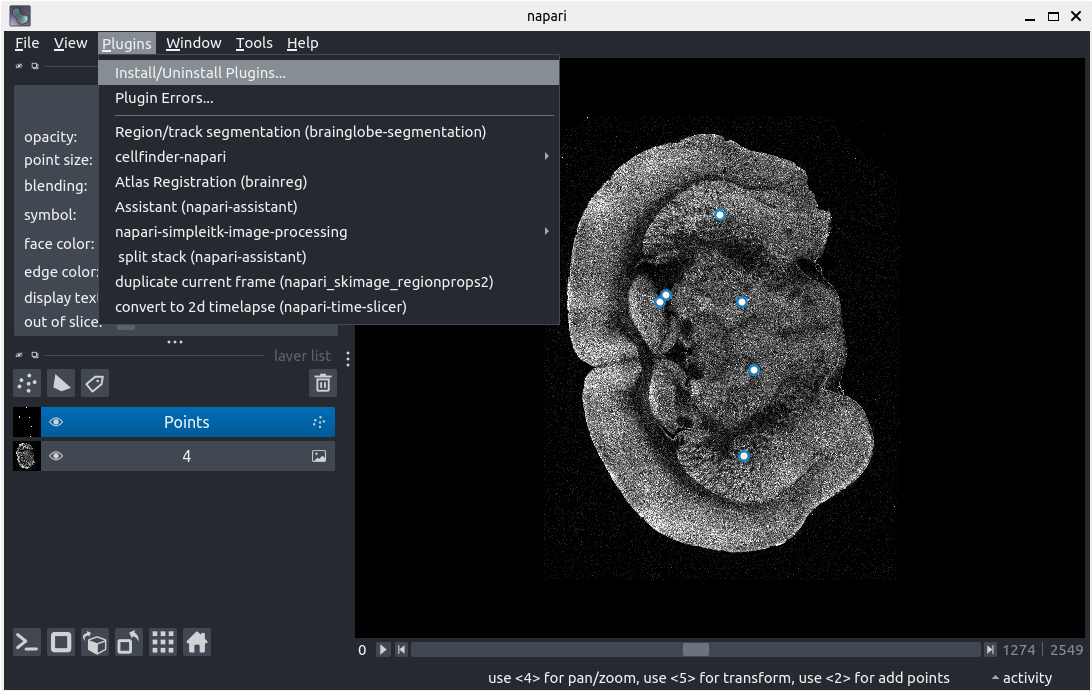
Plugin installation
- Navigate to
Plugins > Install/Uninstall plugins... - Search for
brainrender-napari - Click the
Installbutton - Close the pop-up notification
- Restart
napari - Open
Plugins > Brainrender (brainrender-napari) - Explore the Brainrender plugin
Plugin installation
After following along, your screen should look something like:

Plugin installation

Summary
- You can open, view, edit and save layers in
napari - You can run Python commands with the
napariconsole - You can install and open plugins
Registration & segmentation
Cell detection
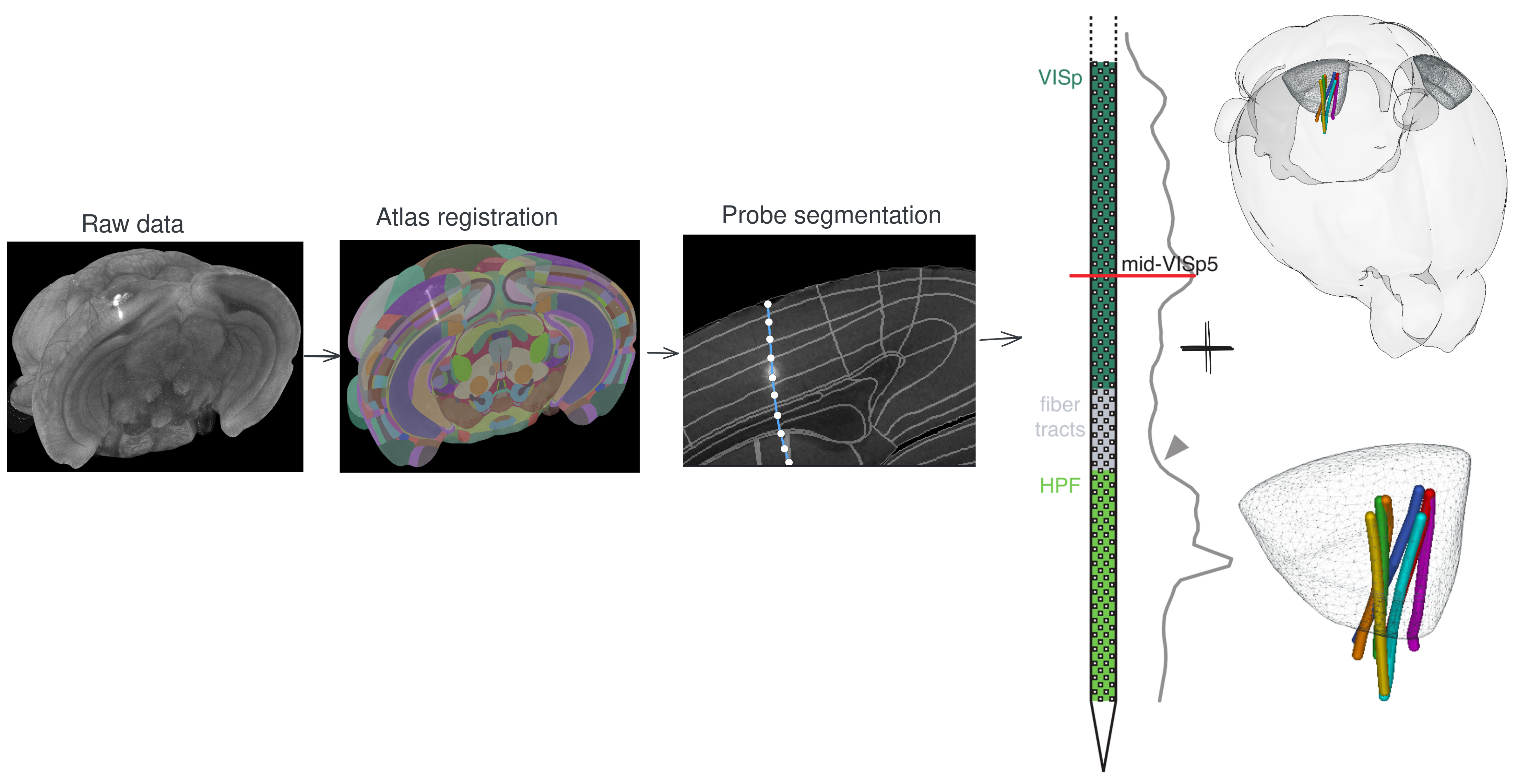
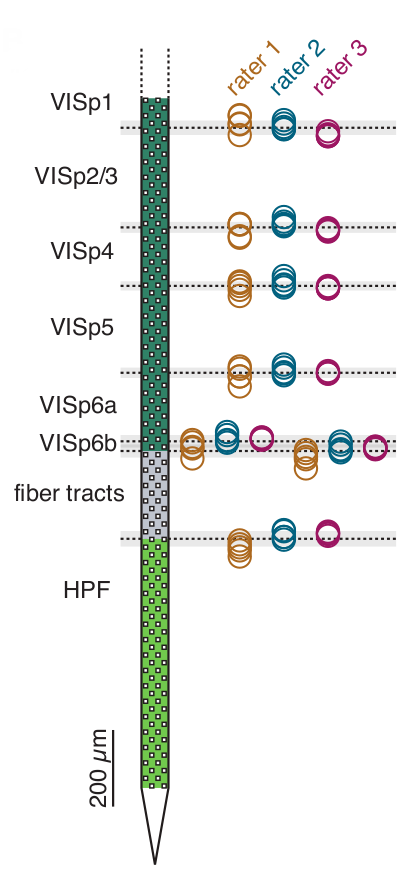
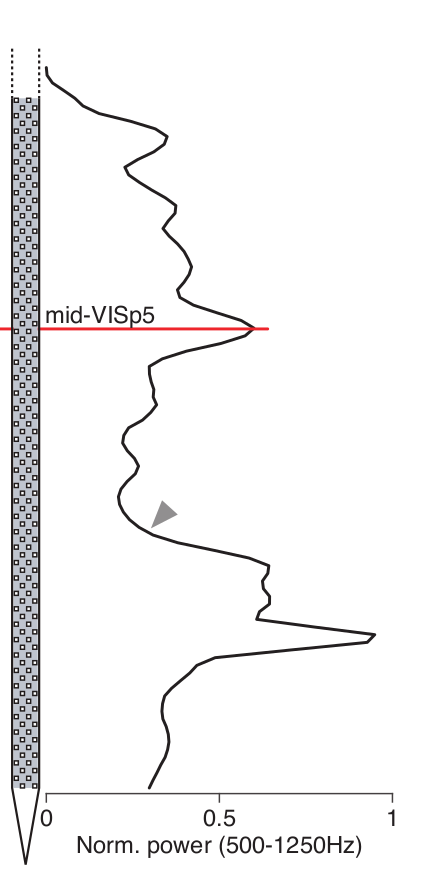
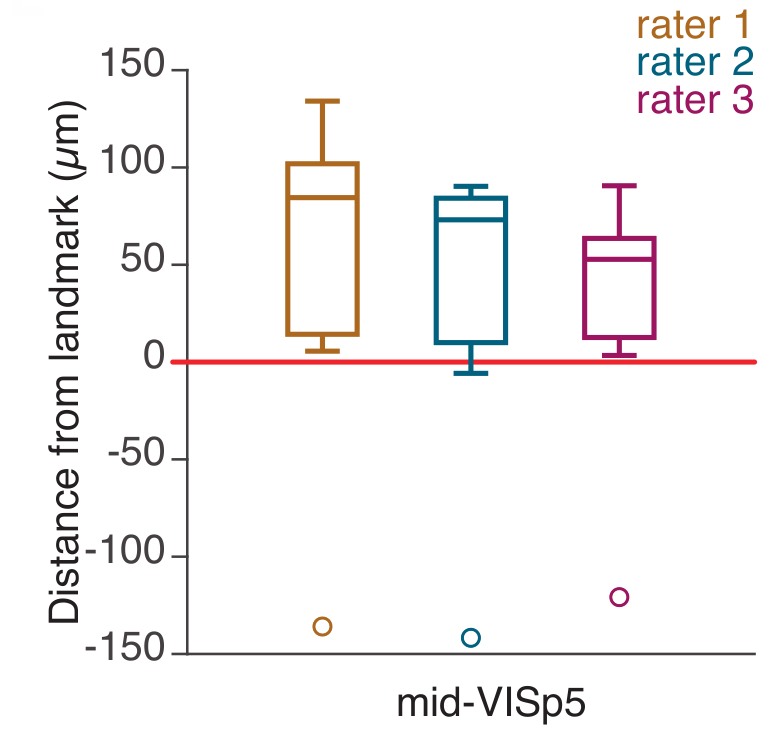
Silicon probe tracing
Bulk fluorescence mapping
BrainGlobe & your software
BrainGlobe & your software
- Not just standalone tools
- Python APIs to help you build your own software
- Easy compatibility with other packages
- Released tools
Example script
- Find all hippocampal regions (
bg-atlasapi) - Display them in napari (
brainrender-napari)
Example script (demo)
import napari
from bg_atlasapi import BrainGlobeAtlas
from brainrender_napari.napari_atlas_representation import NapariAtlasRepresentation
# setup a napari viewer and a brainglobe atlas
viewer = napari.viewer.Viewer()
viewer.dims.ndisplay = 3 # set to 3d mode
atlas = BrainGlobeAtlas("allen_mouse_100um")
# find all hippocampal regions
hip_id = 1080 # the id of the hippocampus is 1080
hip_regions = [
region["acronym"]
for region in atlas.structures_list
if hip_id in region["structure_id_path"]
]
# make a representation of the brainglobe atlas in napari
napari_atlas = NapariAtlasRepresentation(atlas, viewer)
# add all hippocampal regions to the napari viewer
for hip_region in hip_regions:
napari_atlas.add_structure_to_viewer(hip_region)
# add the whole brain mesh as a help for orientation
napari_atlas.add_structure_to_viewer("root")
# run this script
if __name__ == "__main__":
napari.run()Wrap up
Resources
You are welcome to contribute to BrainGlobe - get in touch anytime and we will support you!
Q+A
Feedback
Please give us some feedback on this pilot course.
Thank you!
SWC | 2023-12-06