Cellfinder version 1.3.0 is released!#
We are excited to announce that a new version of cellfinder has been released.
Main updates#
This update brings a significant change to the backend of
cellfinder, as we have switched from TensorFlow to PyTorch. This change allowscellfinderto support python versions 3.11+, and simplifies the installation process. The newcellfinderversion maintains the same classification accuracy. Models trained using previous versions ofcellfinderwill continue to work with the new version.The default batch size used for detection has been increased to 64, which improves classification speed by approximately 40% on most systems. The batch size used for detection can now also be adjusted in the
napariplugin.
What do I need to do?#
We recommend using a fresh conda environment to simplify the update. For GPU support, please follow the installation instructions in the documentation.
conda create -n cellfinder -c conda-forge python=3.11
conda activate cellfinder
pip install cellfinder
You can also update an existing installation of cellfinder using pip:
pip install --upgrade cellfinder
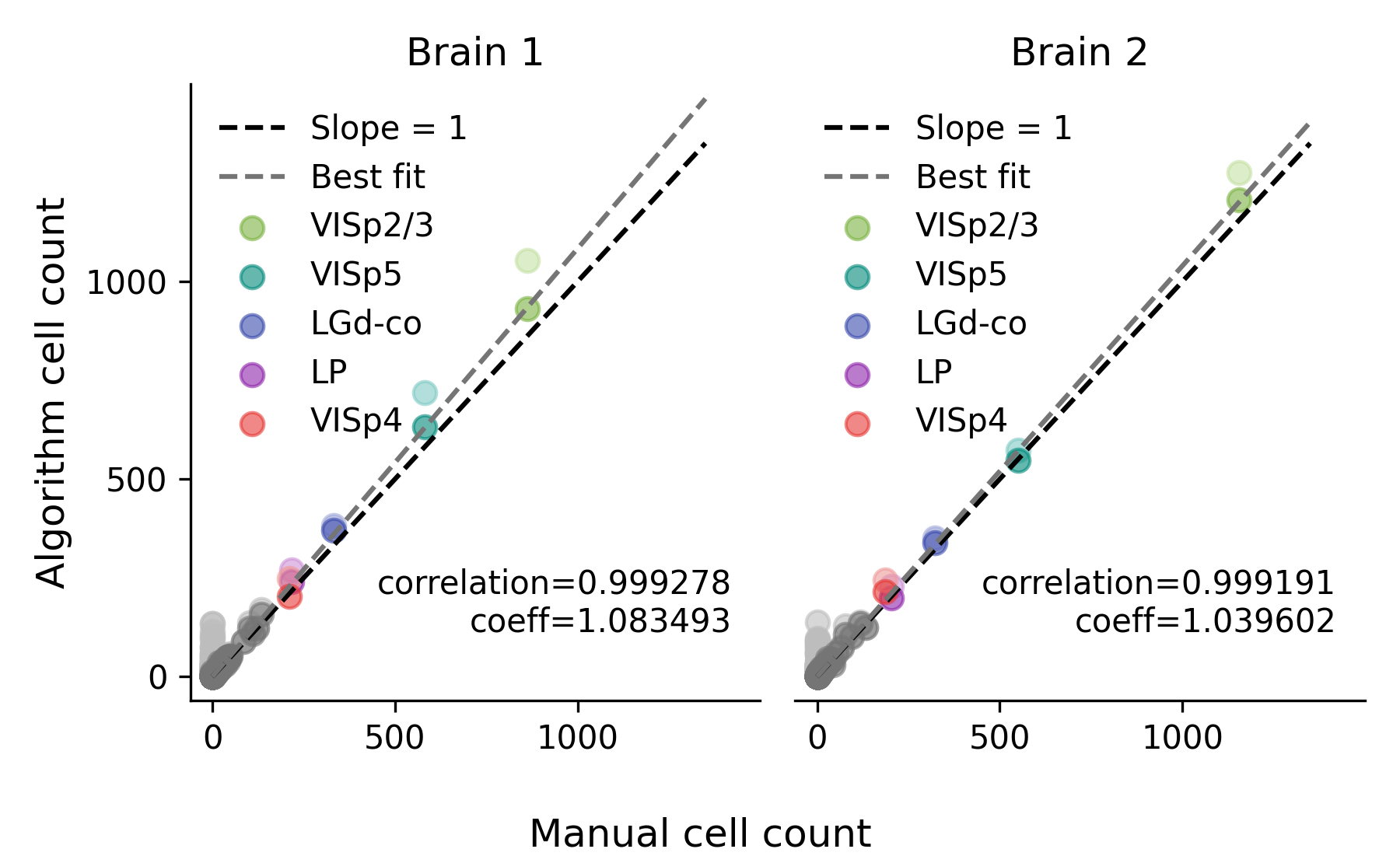
Classification performance#
The classification performance between the two versions is comparable. Below is a comparison of the performance between the two versions using data from the cellfinder paper. Running cellfinder with a PyTorch backend results in a comparable Pearson correlation and slightly improved linear best-fit slope (labelled as “coeff” in the plot) when comparing to manual cell counts. For more details on how the plots were generated, see the cellfinder paper.
TensorFlow backend#
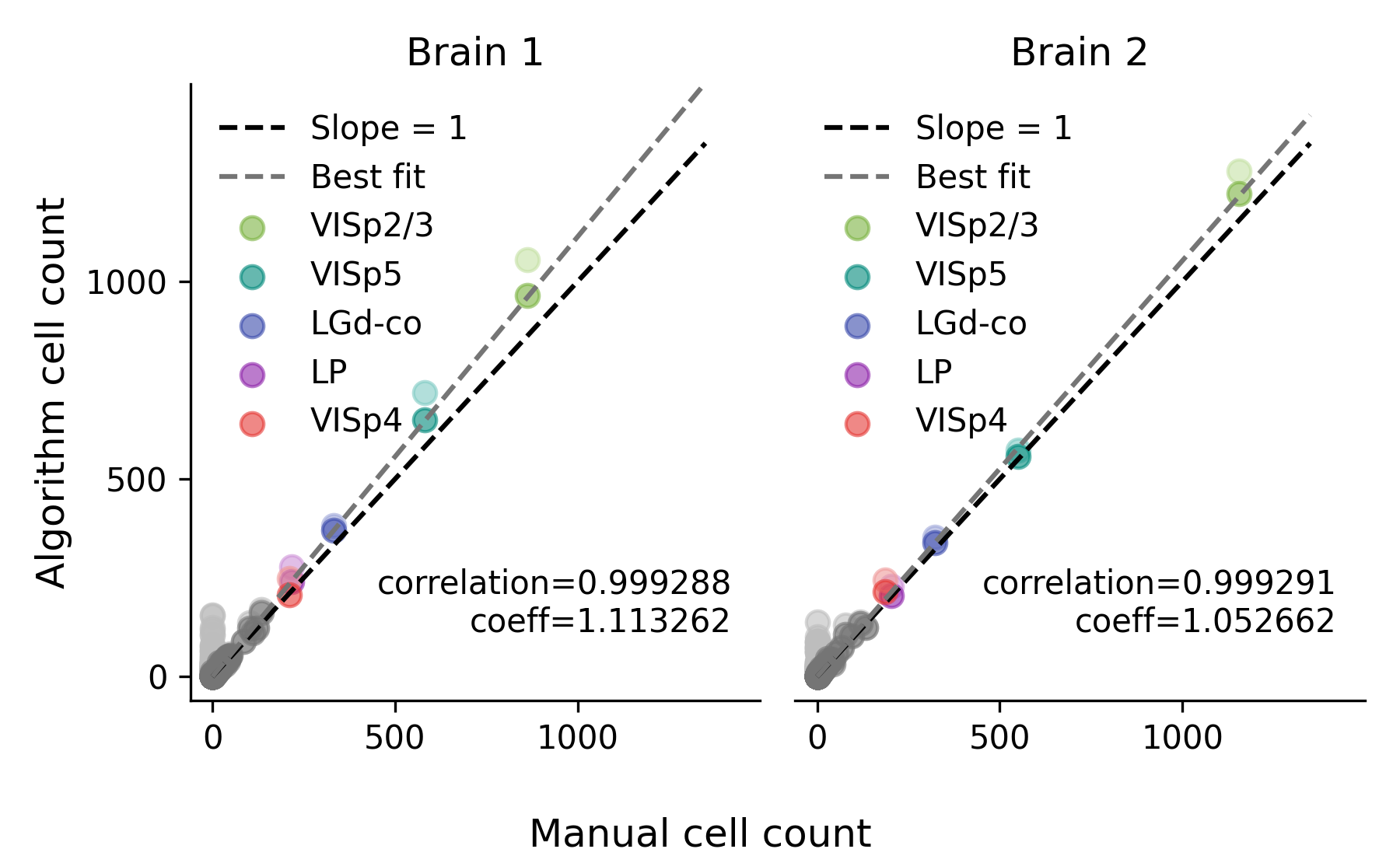
PyTorch backend#