New mouse brain atlas based on gene expression#
The CArea atlas has been added to BrainGlobe. Regions in this atlas are delineated based on clustering 3D volumes of gene expression, finding voxels with similar “gene expression fingerprints”. Data from over 1000 mice was used to generate both volumes of gene expression, and an average Nissl template. This template is a slightly improved and updated version the CCFv3BBP template (also in BrainGlobe as ccfv3augmented_mouse).
In the future, atlases regions may be defined automatically based on cell type, pathology, or other properties which can be mapped to a template. A paper describing this atlas and the accompanying gene expression dataset is now available on bioRxiv. The ISH data is originally sourced from the Allen Brain Atlas ISH dataset. The reconstructed gene volumes have a web app for searching and interacting with them.
Atlas name:
carea_mouse_25umResolution: 25 µm isotropic
Template space: Aligned to the CCFv3BBP
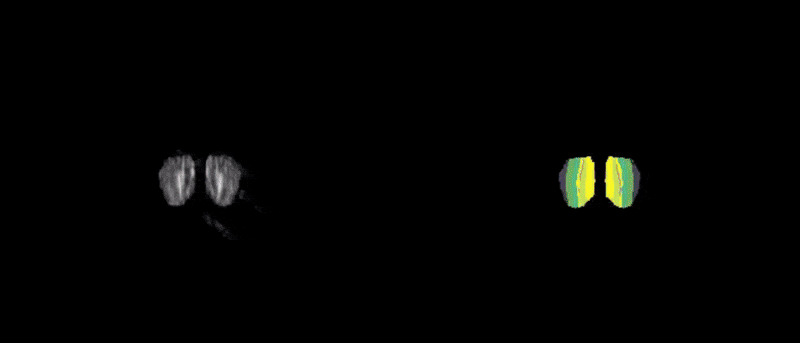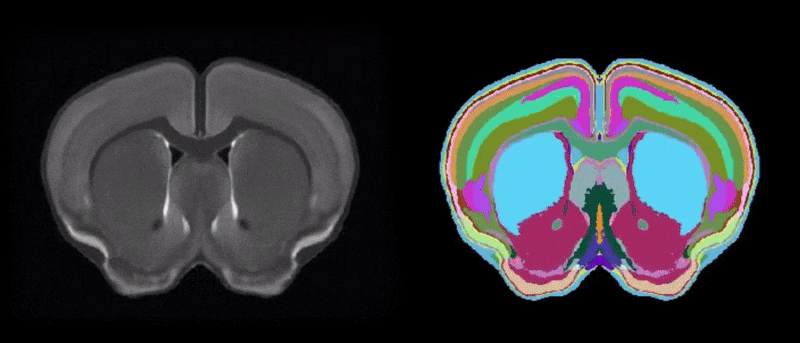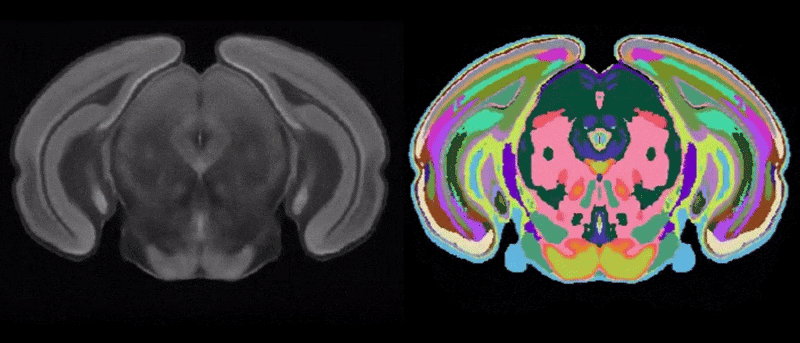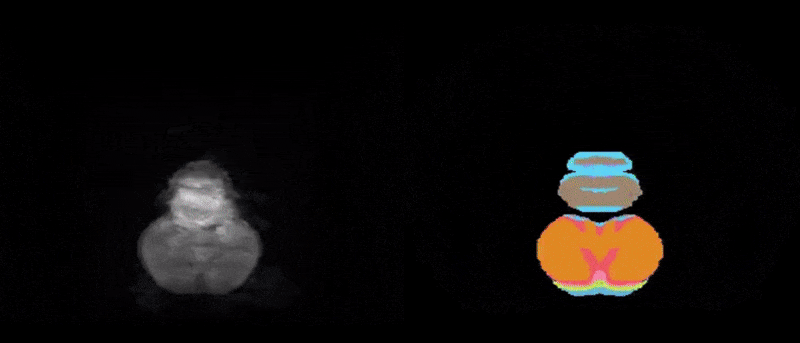
Figure 1. Two-dimensional view of the CArea Mouse atlas: reference (left) and annotations (right)
How do I use the new atlas?#
You can use the CArea mouse atlas like all other BrainGlobe atlases. To visualise the atlas, you could follow the steps below:
Install BrainGlobe (instructions).
Open Napari and follow the steps in our download tutorial for the
carea_mouse_25umatlas.Run
napari -w brainrender-napariand visualise the different parts of the atlas as described in our visualisation tutorial.
The end result will look something like Figure 2.
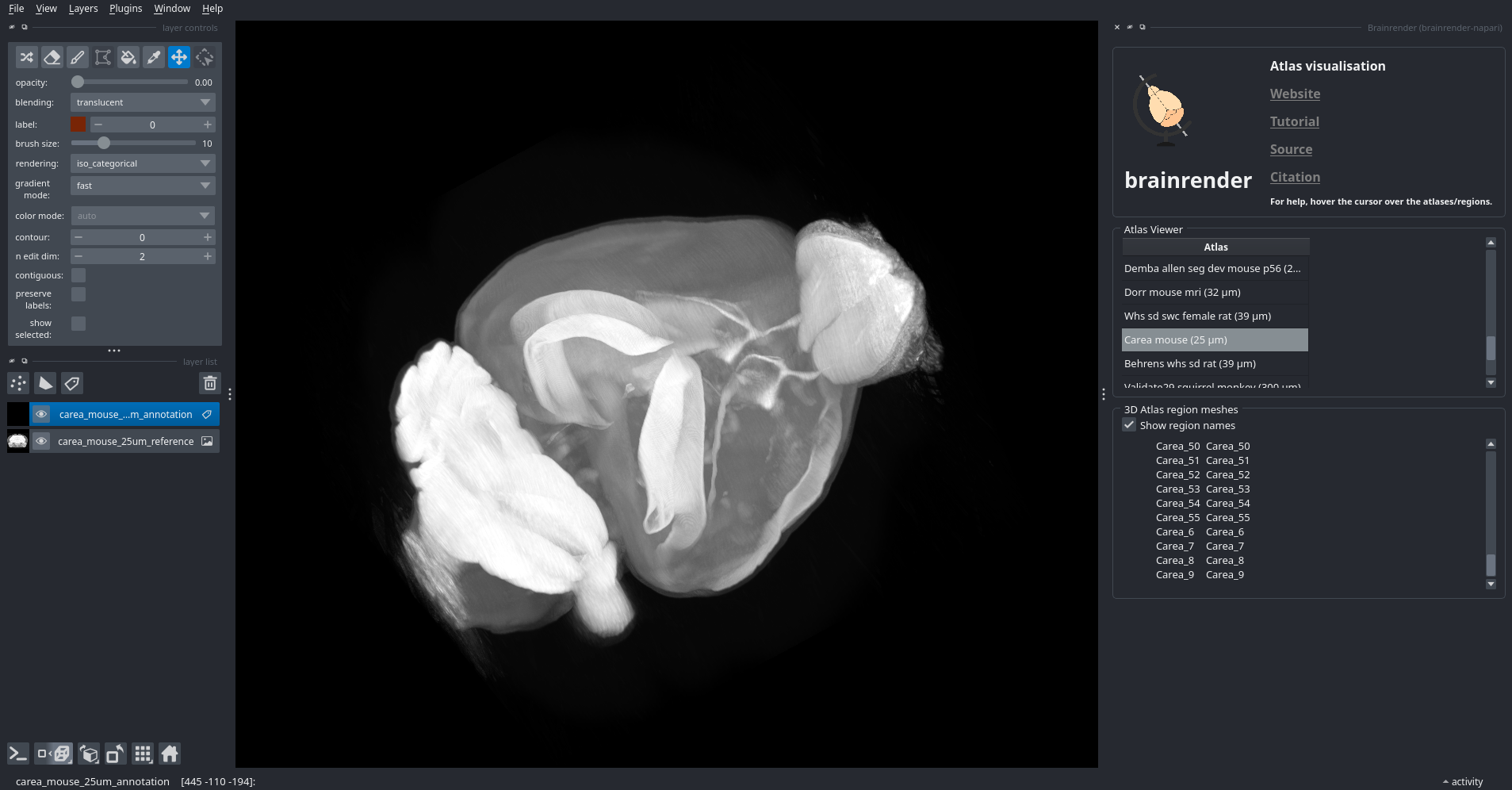 Figure 2. CArea atlas visualised with
Figure 2. CArea atlas visualised with brainrender-napari.
Why are we adding new atlases?#
A fundamental aim of the BrainGlobe project is to make various brain atlases easily accessible by users across the globe. If you would like to get involved with a similar project, please get in touch.
