cellfinder#
cellfinder is software for automated 3D cell detection in very large 3D images (e.g., serial two-photon or lightsheet volumes of whole mouse brains).
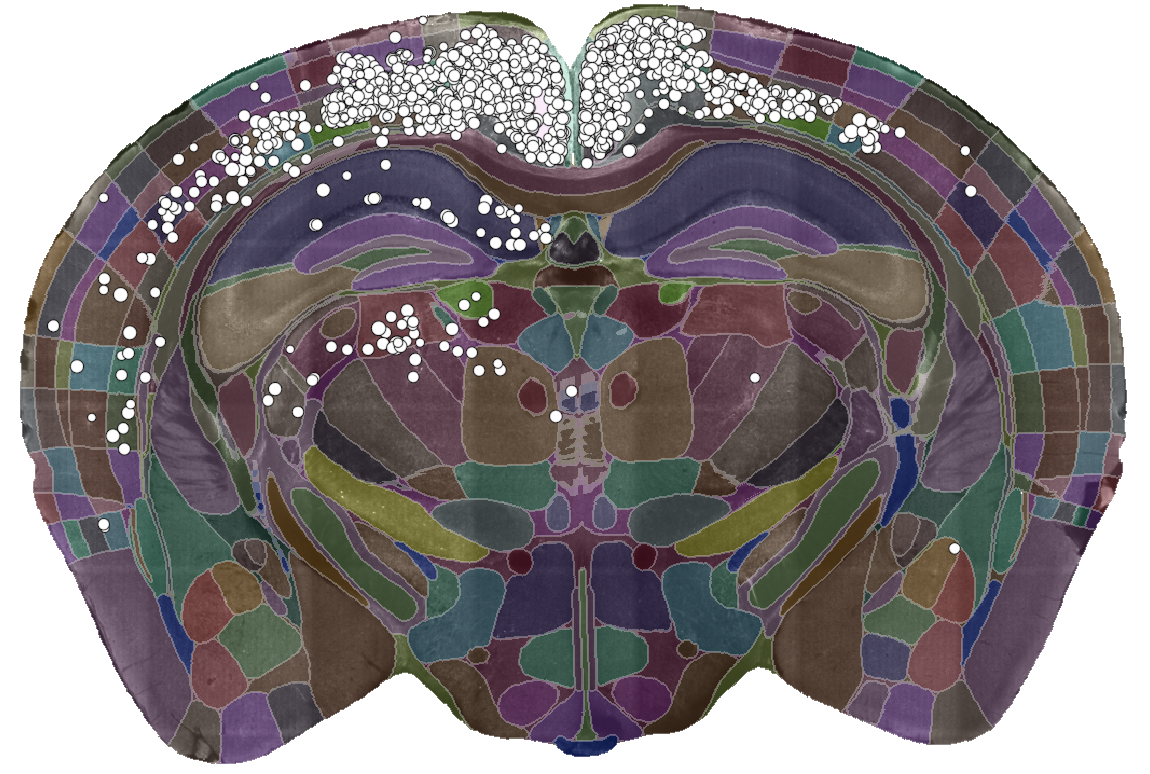 Detected labelled cells, overlaid on a segmented coronal brain section
Detected labelled cells, overlaid on a segmented coronal brain section
Ways to use cellfinder#
cellfinder can be used in three ways, each with different user interfaces and different aims.
cellfinder.core#
cellfinder.core is a Python submodule implementing the core algorithm for efficient cell detection in large images.
The submodule exists to allow developers to implement the algorithm in their own software.
cellfinder napari plugin#
This is a thin wrapper around the cellfinder.core submodule and aims to:
Provide the cell detection algorithm in a user-friendly form
Allow the cell detection algorithm to be chained together with other tools in the napari ecosystem
Allow easier parameter optimisation for users of the other cellfinder tools.
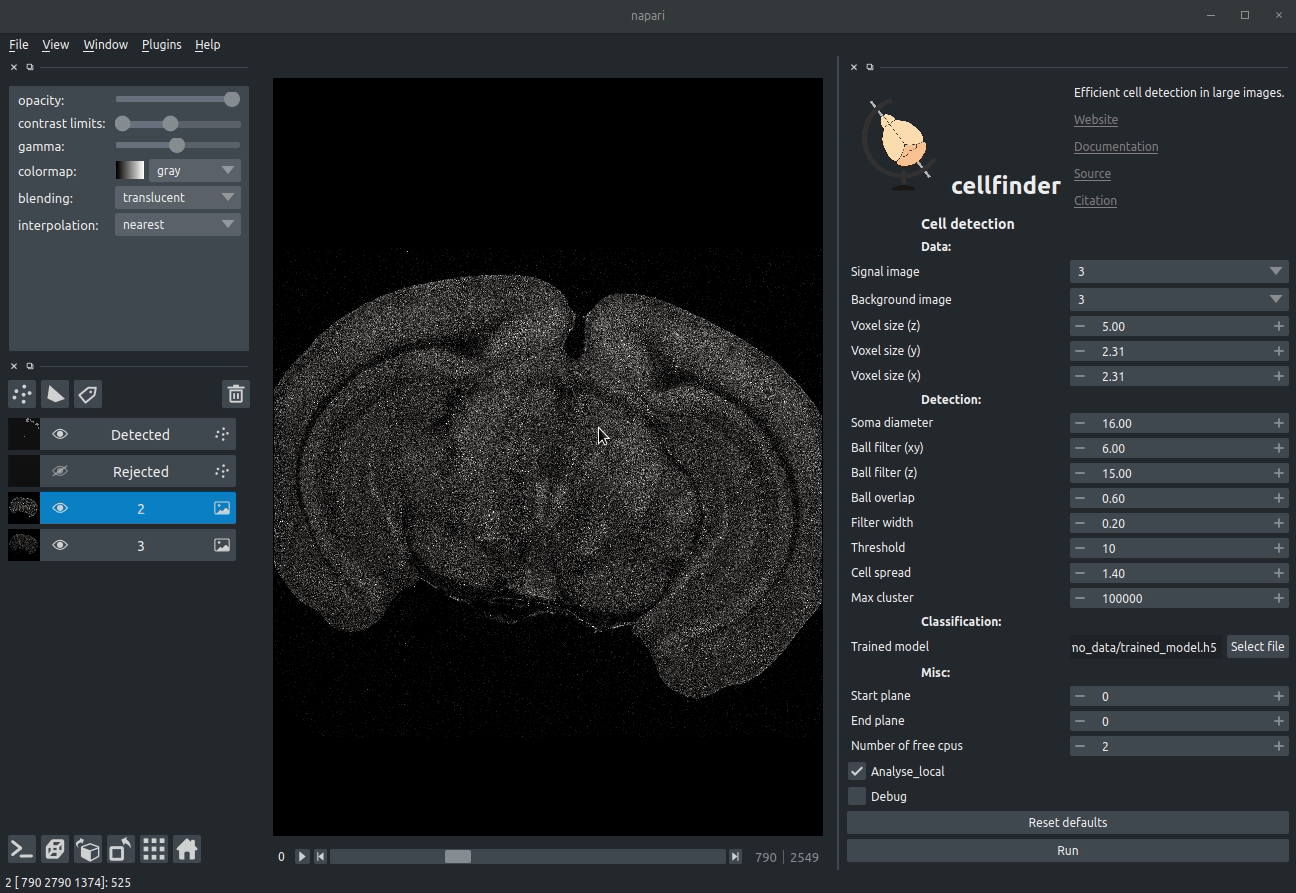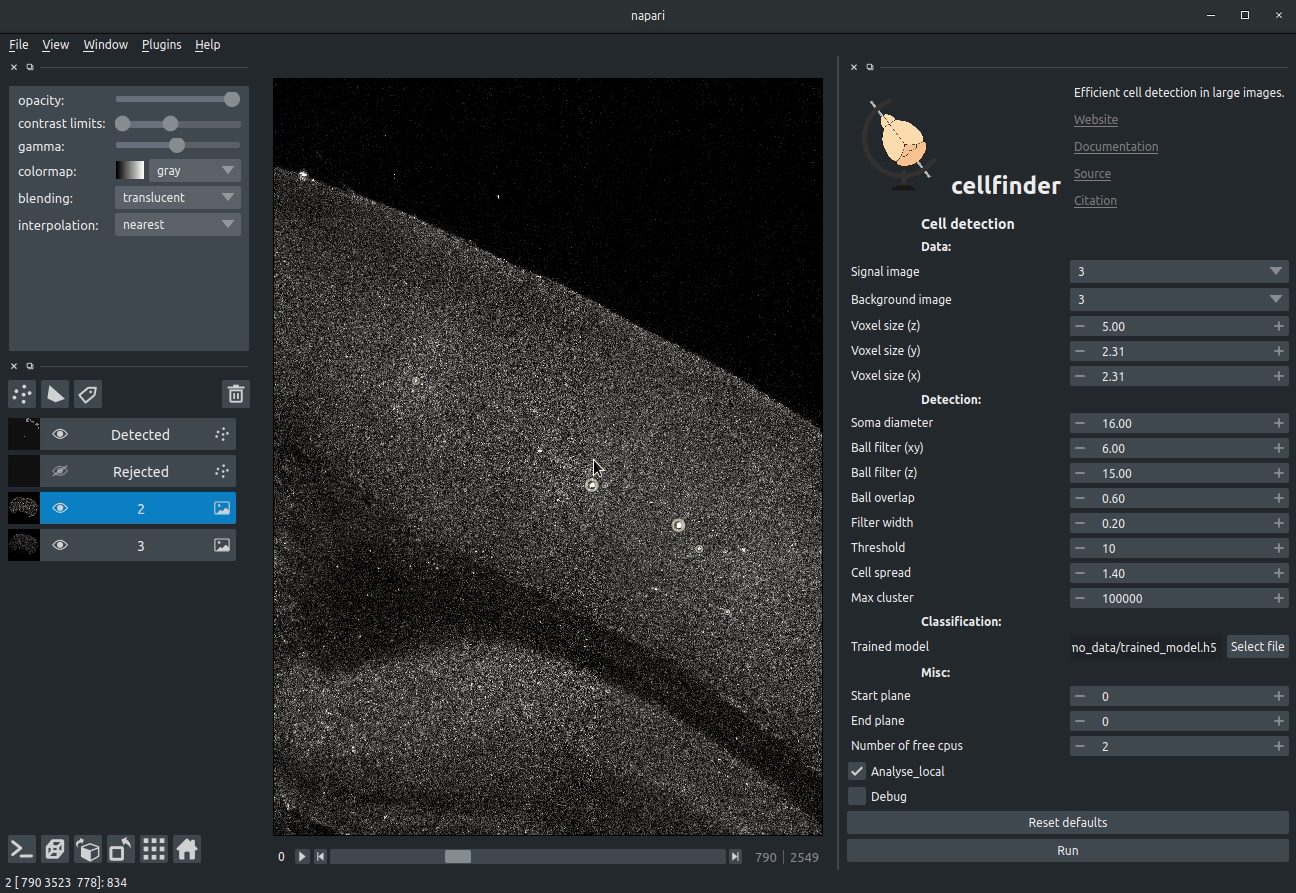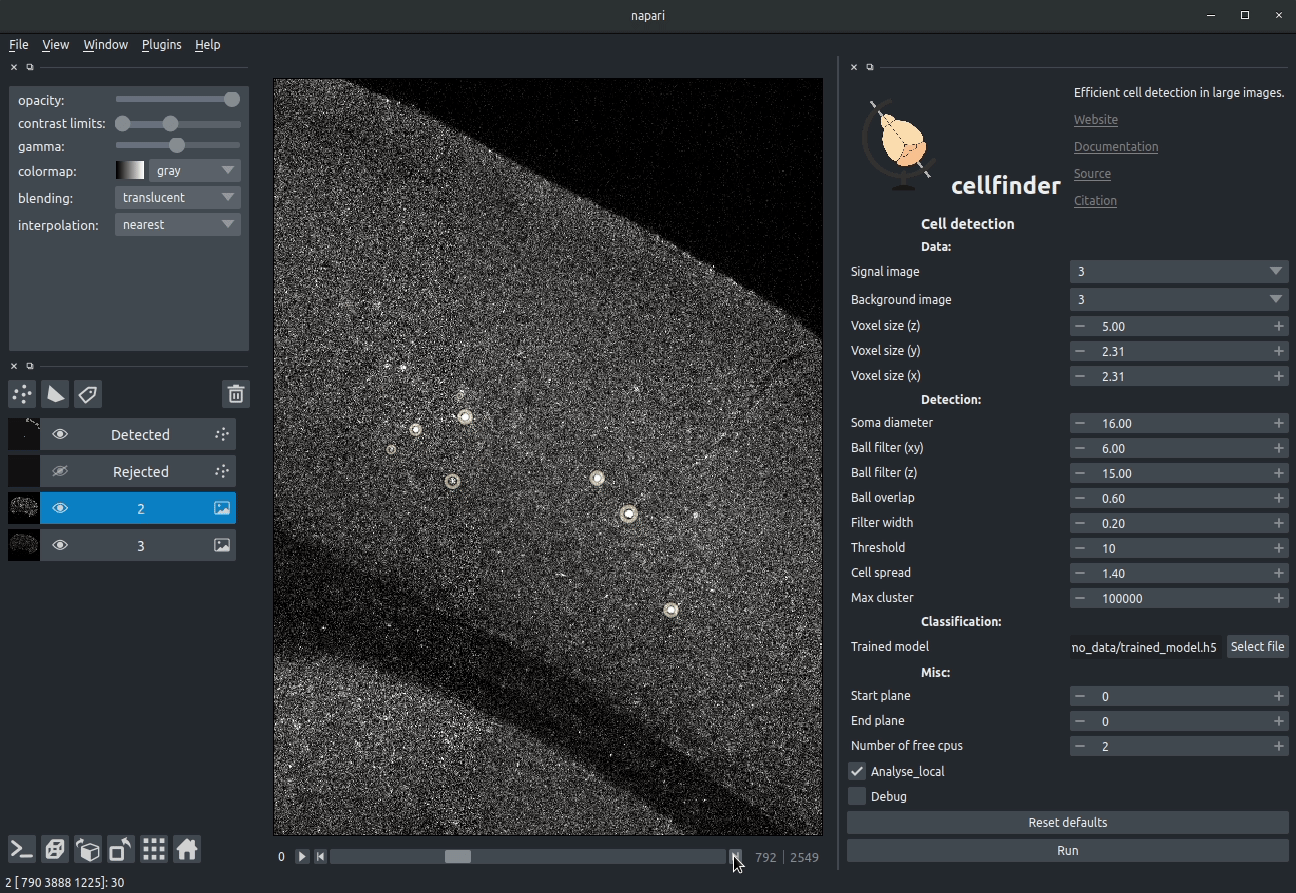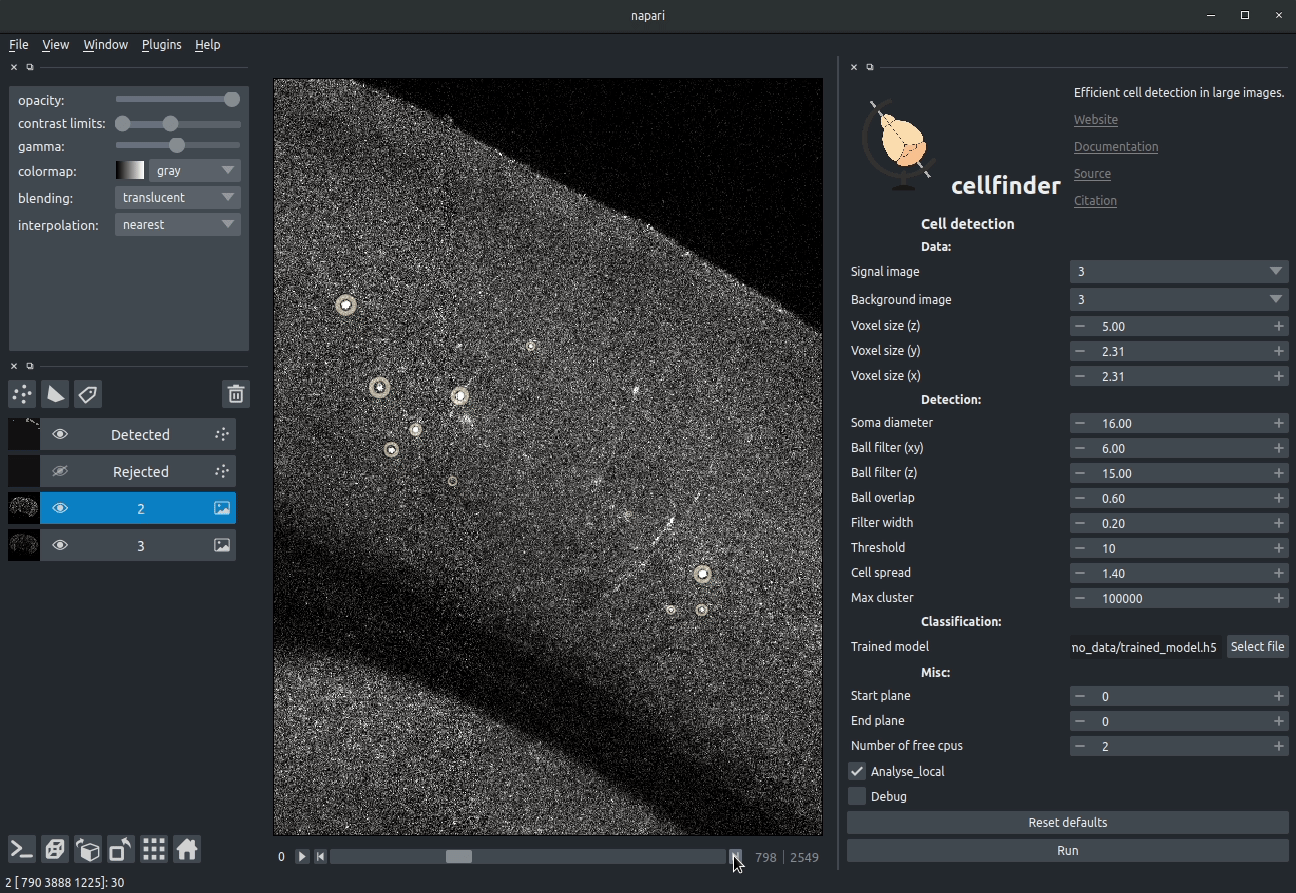 Visualising detected cells in the cellfinder napari plugin
Visualising detected cells in the cellfinder napari plugin
brainmapper command-line tool#
The brainmapper command-line tool exists to combine the cellfinder.core cell detection algorithm and brainreg.
See the documentation for brainglobe-workflows for more information.
Installation#
User guide#
Troubleshooting#
Citing cellfinder#
If you find cellfinder useful, and use it in your research, please cite the paper outlining the cell detection algorithm:
Tyson, A. L., Rousseau, C. V., Niedworok, C. J., Keshavarzi, S., Tsitoura, C., Cossell, L., Strom, M. and Margrie, T. W. (2021) “A deep learning algorithm for 3D cell detection in whole mouse brain image datasets’ PLOS Computational Biology, 17(5), e1009074 https://doi.org/10.1371/journal.pcbi.1009074
